Deep neural network-based prediction of RNA aptamers targeting E6 protein of high-risk human papilloma virus
Main Article Content
Abstract
Background: The Human papilloma virus is the primary cause of cervical cancer. The virus integrates with the human genome to produce the E6 oncoprotein. Therefore, the E6 oncoprotein is a crucial molecular target for cancer progression or treatment. The development of aptamers is beneficial for interacting with the target protein and serves as a new strategy for detection or delivery systems.
Objectives: We aim to explore the candidate aptamer sequence against E6 oncoprotein using a computational-based method.
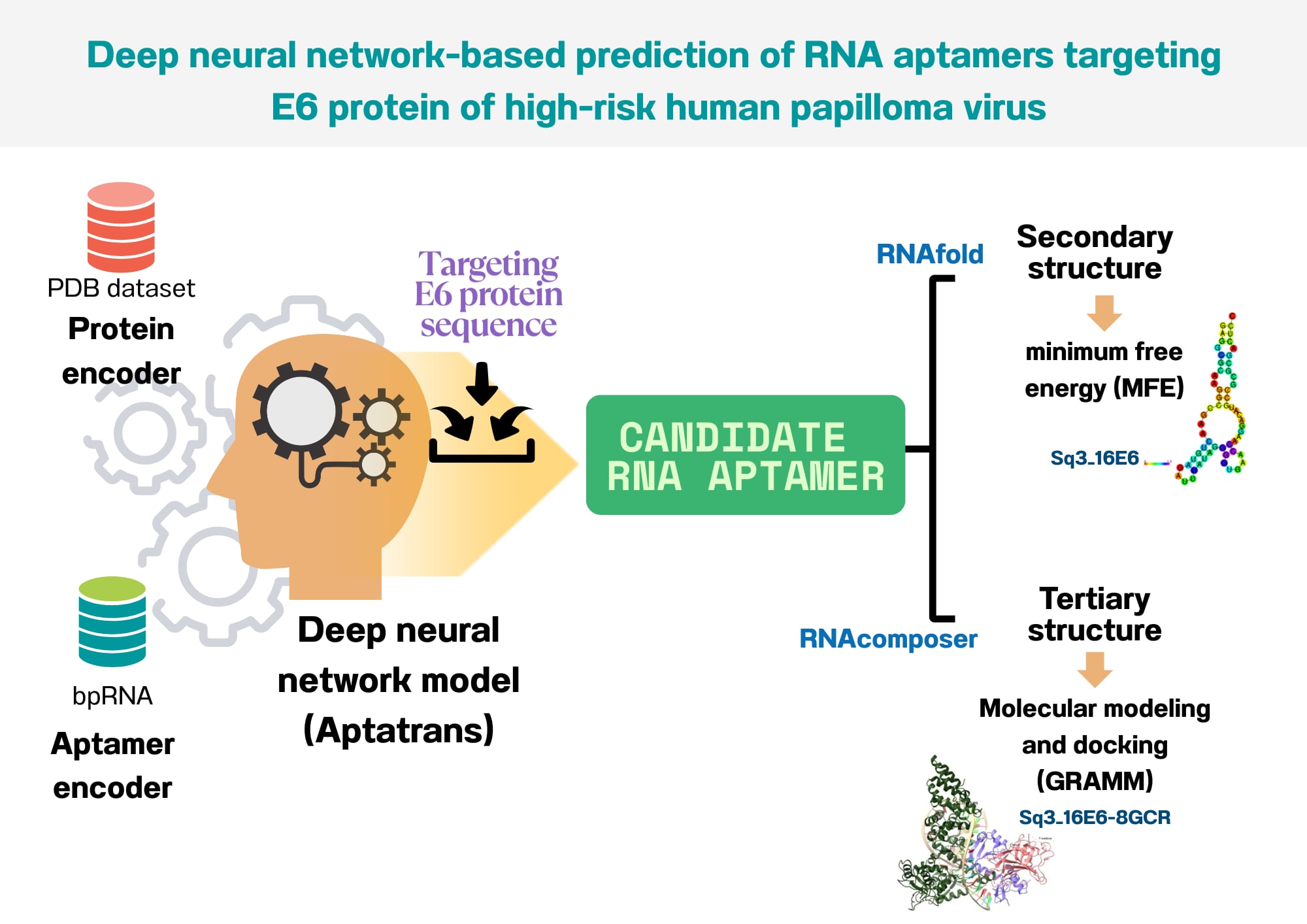
Materials and methods: This study designed the candidate aptamer against the target protein based on computational approaches using the AptaTrans pipeline. After obtaining the candidate aptamer sequences, the minimum free energy was calculated using the RNAfold web server. The tertiary structure was then generated using RNAComposer. Next, the molecular docking score was acquired from the GRAMM web server.
Results: The aptamer sequences with the best stability, as indicated by minimum free energy (MFE), are Sq3_16E6, Sq3_Actn, and Sq3_18E6, respectively. The aptamer sequences of Sq3_16E6 and Sq2_18E6 showed potential interactions with 8GCR and 6SJV, respectively.
Conclusion: Sq3_16E6 and Sq2_18E6 are appropriate for the development of the detection of the E6 protein in cervical swabs. Further investigation should be performed.
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Personal views expressed by the contributors in their articles are not necessarily those of the Journal of Associated Medical Sciences, Faculty of Associated Medical Sciences, Chiang Mai University.
References
Woodman CBJ, Collins SI, Young LS. The natural history of cervical HPV infection: unresolved issues. Nat Rev Cancer. 2007; 7(1): 11-22. doi: 10.1038/nrc2050.
Ploysawang P, Rojanamatin J, Prapakorn S, Jamsri P, Pangmuang P, Seeda K, et al. National cervical cancer screening in Thailand. Asian Pac J Cancer Prev. 2021; 22(1): 25-30. doi: 10.31557/APJCP.2021.22.1.25.
Falcaro M, Castañon A, Ndlela B, Checchi M, Soldan K, Lopez-Bernal J, et al. The effects of the national HPV vaccination programme in England, UK, on cervical cancer and grade 3 cervical intraepithelial neoplasia incidence: a register-based observational study. The Lancet. 2021; 398(10316): 2084-92. doi: 10.1016/S0140-6736(21)02178-4.
Huh WK, Ault KA, Chelmow D, Davey DD, Goulart RA, Garcia FAR, et al. Use of primary high-risk human papillomavirus testing for cervical cancer screening: Interim clinical guidance. Obstet Gynecol. 2015; 125(2): 330-7. doi: 10.1097/AOG.0000000000000669.
Derbie A, Mekonnen D, Woldeamanuel Y, Van Ostade X, Abebe T. HPV E6/E7 mRNA test for the detection of high grade cervical intraepithelial neoplasia (CIN2+):
a systematic review. Infect Agent Cancer. 2020; 15(1): 9. doi: 10.1186/s13027-020-0278-x.
Downham L, Jaafar I, Rol ML, Nyawira Nyaga V, Valls J, Baena A, et al. Accuracy of HPV E6/E7 oncoprotein tests to detect high-grade cervical lesions: a systematic literature review and meta-analysis. Br J Cancer. 2024; 130(4): 517-25. doi: 10.1038/s41416-023-02490-w.
Schweizer J, Lu PS, Mahoney CW, Berard-Bergery M, Ho M, Ramasamy V, et al. Feasibility Study of a Human Papillomavirus E6 Oncoprotein Test for Diagnosis of Cervical Precancer and Cancer. J Clin Microbiol. 2010; 48(12): 4646-8. doi: 10.1128/JCM.01315-10.
Tan S, G.E. De Vries E, G.J. Van Der Zee A, De Jong S. Anticancer drugs aimed at E6 and E7 activity in HPV-positive cervical cancer. Curr Cancer Drug Targets. 2012; 12(2): 170-84. doi: 10.2174/156800912799095135.
Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990; 346(6287): 818-22. doi: 10.1038/346818a0.
Shin I, Kang K, Kim J, Sel S, Choi J, Lee JW, et al. AptaTrans: a deep neural network for predicting aptamer-protein interaction using pretrained encoders. BMC Bioinformatics. 2023; 24(1): 447. doi: 10.1186/s12859-023-05577-6.
Lee G, Jang GH, Kang HY, Song G. Predicting aptamer sequences that interact with target proteins using an aptamer-protein interaction classifier and a Monte Carlo tree search approach. Nebel JC, editor. PLOS ONE. 2021; 16(6): e0253760. doi: 10.1371/journal.pone.0253760.
Danaee P, Rouches M, Wiley M, Deng D, Huang L, Hendrix D. bpRNA: large-scale automated annotation and analysis of RNA secondary structure. Nucleic Acids Res. 2018; 46(11): 5381-94. doi: 10.1093/nar/gky285.
Berman HM. The protein data bank. Nucleic Acids Res. 2000; 28(1): 235-42. doi: 10.1093/nar/28.1.235.
Khabbazian M, Jabbari H. AI-powered aptamer generation. Nat Comput Sci. 2022; 2(6): 356-7. doi: 10.1038/s43588-022-00253-w.
Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov. 2010; 9(7): 537-50. doi: 10.1038/nrd3141.
Muhammad AM, Zari A, Alsubhi NH, Al-Zahrani MH, Alghamdi RA, Labib MM. Novel design of RNA aptamers as cancer inhibitors and diagnosis targeting the tyrosine kinase domain of the NT-3 growth factor receptor using a computational sequence-based approach. Molecules. 2022; 27(14): 4518. doi: 10.3390/molecules27144518.
Bavi R, Liu Z, Han Z, Zhang H, Gu Y. In silico designed RNA aptamer against epithelial cell adhesion molecule for cancer cell imaging. Biochem Biophys Res Commun. 2019; 509(4): 937-42. doi: 10.1016/j.bbrc.2019.01.028.
Navien TN, Thevendran R, Hamdani HY, Tang TH, Citartan M. In silico molecular docking in DNA aptamer development. Biochimie. 2021; 180:5 4-67. doi: 10.1016/j.biochi.2020.10.005.
De Araújo NS, Moreira ADS, Abreu RDS, Junior VV, Antunes D, Mendonça JB, et al. Aptamer-based recognition of breast tumor cells: A new rra for breast cancer diagnosis. Int J Mol Sci. 2024; 25(2): 840. doi: 10.3390/ijms25020840.
Singh A, Copeland MM, Kundrotas PJ, Vakser IA. GRAMM web server for protein docking. In: Gore M, Jagtap UB, Editors. Computational rug Discovery and Design (Methods in Molecular Biology; Vol. 2714). New York, NY: Springer US, pp 101-12. doi: 10.1007/978-1-0716-3441-7_5.
Ji C, Wei J, Zhang L, Hou X, Tan J, Yuan Q, et al. Aptamer–protein interactions: From regulation to biomolecular detection. Chem Rev. 2023; 123(22): 12471-506. doi: 10.1021/acs.chemrev.3c00377.