MALDI-TOF mass spectrometry in transfusion medicine
Main Article Content
Abstract
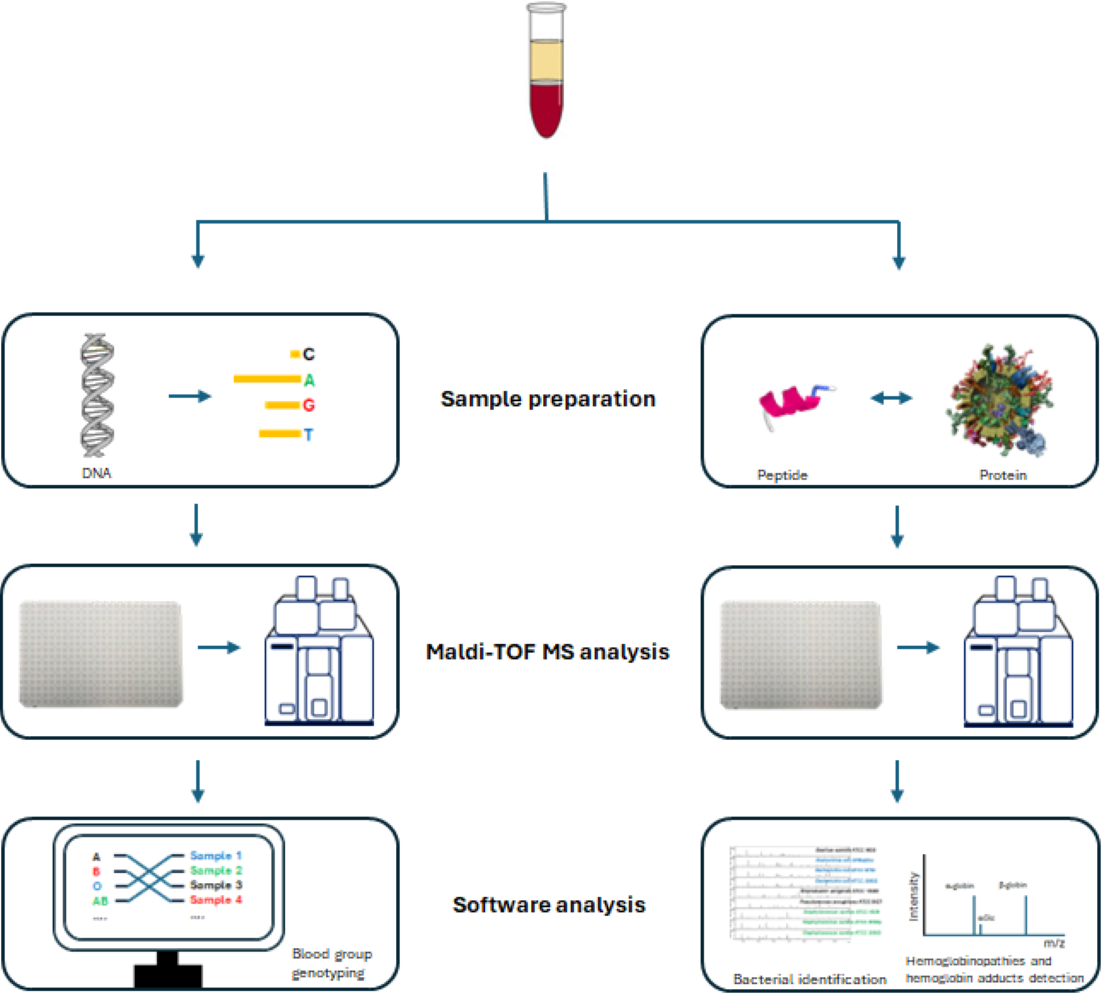
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) is a valuable tool in clinical research. In transfusion medicine, DNA-based genotyping is increasingly replacing serological methods for precise blood group determination. MALDI-TOF MS platforms for SNP genotyping have been rigorously developed and validated against serological techniques. MALDITOF MS offers high-throughput capacity, exceptional multiplexing, and direct detection, resulting in high accuracy and flexibility. The ability to perform blood group genotyping, bacterial identification via peptide barcoding, and detection of hemoglobinopathies and hemoglobin adducts in both donors and recipients makes MALDI-TOF MS a promising technology for resolving complex blood typing cases and enhancing clinical utility in transfusion medicine.
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Personal views expressed by the contributors in their articles are not necessarily those of the Journal of Associated Medical Sciences, Faculty of Associated Medical Sciences, Chiang Mai University.
References
Storry JR, Clausen FB, Castilho L, Chen Q, Daniels G, Denomme G, et al. International Society of Blood Transfusion Working Party on Red Cell Immunogenetics and Blood Group Terminology: Report of the Dubai, Copenhagen and Toronto meetings. Vox Sang. 2019; 114(1): 95-102. doi: 10.1111/vox.12717.
Table of blood group systems v11.2 https://www.isbtweb.org/resource/tableofbloodgroupsystems.html Date: 22-JAN-2024
Siebert PD, Fukuda M. Molecular cloning of a human glycophorin B cDNA: Nucleotide sequence and genomic relationship to glycophorin A. Proc Natl Acad Sci USA. 1987; 84: 6735-9. doi: 10.1073/pnas.84.19. 6735.
Yamamoto F, Clausen H, White T, Marken J, Hakomori S. Molecular genetic basis of the histo-blood group ABO system. Nature. 1990; 345(6272): 229-33. doi:10.1038/345229a0.
Daniels G. Human Blood Groups. Oxford: Wiley-Blackwell; 2013.
Avent ND. Recombinant technology in transfusion medicine. Curr Pharm Biotechnol. 2000; 1(2): 117-35. doi: 10.2174/1389201003378951.
Dzik WH. Molecular diagnostics in transfusion medicine:the best is yet to come. Transfusion. 1995; 35(3): 183-5. doi: 10.1046/j.1537-2995.1995.35395184271.x.
Westhoff CM, Sloan SR. Molecular genotyping in transfusion medicine. Clin Chem. 2008; 54(12): 1948-50. doi: 10.1373/clinchem.2008.116038.
Marvin LF, Roberts MA, Fay LB. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in clinical chemistry. Clin Chim Acta. 2003; 337(1-2):11-21. doi: 10.1016/j.cccn.2003.08.008.
Calderaro A, Chezzi C. MALDI-TOF MS: A reliable tool in the real life of the clinical microbiology laboratory. Microorganisms. 2024 Feb 3;12(2):322. doi: 10.3390/microorganisms12020322.
Sousa P, Silva L, Luís C, Câmara JS, Perestrelo R.MALDI-TOF MS: A promising analytical approach to cancer diagnostics and monitoring. Separations. 2023; 10(8): 453. doi.org/10.3390/separations10080453.
Cho YT, Su H, Huang TL, Chen HC, Wu WJ, Wu PC, et al. Matrix-assisted laser desorption ionization/timeof-flight mass spectrometry for clinical diagnosis. Clin Chim Acta. 2013; 415: 266-75. doi: 10.1016/j.cca.2012.10.032.
Perry WJ, Patterson NH, Prentice BM, Neumann EK, Caprioli RM, Spraggins JM. Uncovering matrix effects on lipid analyses in MALDI imaging mass spectrometry experiments. J Mass Spectrom. 2020; 55(4): e4491. doi: 10.1002/jms.4491.
Pusch W, Kostrzewa M. Application of MALDI-TOF mass spectrometry in screening and diagnostic research. Curr Pharm Des. 2005; 11(20): 2577-91. doi: 10.2174/1381612054546932.
Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007; 5(10): e254. doi: 10.1371/journal.pbio.0050254.
Spengler B, Pan Y, Cotter RJ, Kan LS. Molecular weight determination of underivatized oligodeoxyribonucleotides by positive-ion matrix-assisted ultraviolet laser-desorption mass spectrometry. Rapid Commun Mass Spectrom. 1990; 4(4): 99-102. doi:10.1002/rcm.1290040402.
Shahgholi M, Garcia BA, Chiu NH, Heaney PJ, Tang K. Sugar additives for MALDI matrices improve signal allowing the smallest nucleotide change (A:T) in a DNA sequence to be resolved. Nucleic Acids Res. 2001; 29(19): E91. doi: 10.1093/nar/29.19.e91.
Asara JM, Allison J. Enhanced detection of oligonucleotides in UV MALDI MS using the tetraamine spermine as a matrix additive. Anal Chem. 1999; 71(14): 2866-70. doi: 10.1021/ac981406l.
Schuerenberg M, Luebbert C, Eickhoff H, Kalkum M, Lehrach H, Nordhoff E. Prestructured MALDI-MS sample supports. Anal Chem. 2000; 72(15): 3436-42. doi.org/10.1021/ac000092a.
Nordhoff E, Ingendoh A, Cramer R, Overberg A, Stahl B, Karas M, et al. Matrix-assisted laser desorption/ionization mass spectrometry of nucleic acids with wavelengths in the ultraviolet and infrared. Rapid Commun Mass Spectrom. 1992; 6(12): 771-6. doi:10.1002/rcm.1290061212.
Little DP, Braun A, Darnhofer-Demar B, Köster H. Identification of apolipoprotein E polymorphisms using temperature cycled primer oligo base extension and mass spectrometry. Eur J Clin Chem Clin Biochem. 1997; 35(7): 545-8. PMID: 9263732.
Nordhoff E, Ingendoh A, Cramer R, Overberg A, Stahl B, Karas M, et al. Matrix-assisted laser desorption/ionization mass spectrometry of nucleic acids with wavelengths in the ultraviolet and infrared. Rapid Commun Mass Spectrom. 1992 Dec;6(12):771-6. doi:10.1002/rcm.1290061212.
Yang H, Wang H, Wang J, Cai Y, Zhou G, He F, et al. Multiplex single-nucleotide polymorphism genotyping by matrix-assisted laser desorption/ionization timeof-flight mass spectrometry. Anal Biochem. 2003; 314(1): 54-62. doi: 10.1016/s0003-2697(02)00641-3.
Bray MS, Boerwinkle E, Doris PA. High-throughput multiplex SNP genotyping with MALDI-TOF mass spectrometry: practice, problems and promise. Hum Mutat. 2001; 17(4): 296-304. doi: 10.1002/humu.27.
Nakai K, Habano W, Nakai K, Fukushima N, Fujita T, Gurwitz D. Ethnic differences of coronary artery disease-associated SNPs in two Israeli healthy populations using MALDI-TOF mass spectrometry. Life Sci. 2004; 75(8): 1003-10. doi: 10.1016/j.lfs.2004.02.012.
Shi Y, Xiang P, Li L, Shen M. Analysis of 50 SNPs in CYP2D6, CYP2C19, CYP2C9, CYP3A4 and CYP1A2 by MALDI-TOF mass spectrometry in Chinese Han population. Forensic Sci Int. 2011; 207(1-3): 183-7. doi: 10.1016/j.forsciint.2010.10.004.
Xu X, Liu Y, Hu H, Wang J, Cai Y, Xie J, et al. Relationship between cancer stem cell-related SNPs and survival outcomes in patients with primary lung cancer. World J Surg Oncol. 2023; 21(1): 243. doi: 10.1186/s12957-023-03064-z.
Zhang Z, Guo Z, Gan T, Huang S, Shang D. MALDI-TOF MS-based SNP assay used to determine the appropriate antidepression for Chinese patients. J Pharm Biomed Anal. 2025; 252: 116460. doi:
1016/j.jpba.2024.116460.
Roback J D, Comb, M R, Ness P M. (Eds.). AABB Technical Manual. 20th Ed. AABB; 2019.
Hanzlick, R. L. Transfusion reactions: A practical approach. American Society of Clinical Pathology Press; 2015.
Finning K, Bhandari R, Sellers F, Revelli N, Villa MA, Muñiz-Díaz E, et al. Evaluation of red blood cell and platelet antigen genotyping platforms (ID CORE XT/ ID HPA XT) in routine clinical practice. Blood Transfus. 2016; 14(2): 160-7. doi: 10.2450/2015.0124-15.
Bonet Bub C, Castilho L. ID CORE XT as a tool for molecular red blood cell typing. Expert Rev Mol Diagn. 2019; 19(9): 777-83. doi: 10.1080/14737159.2019.1656529.
Boccoz SA, Fouret J, Roche M, Lachuer J, Legras-Lachuer C, Corgier BP, et al. Massively parallel and multiplex blood group genotyping using next-generationsequencing. Clin Biochem. 2018; 60: 71-76. doi: 10.1016/j.clinbiochem.2018.07.010.
Kim TY, Yu H, Phan MT, Jang JH, Cho D. Application of blood group genotyping by next-generation
sequencing in various immunohaematology cases. Transfus Med Hemother. 2021; 49(2): 88-96. doi:
1159/000517565.
Sandler SG, Chen LN, Flegel WA. Serological weak D phenotypes: a review and guidance for interpreting the RhD blood type using the RHD genotype. Br J Haematol. 2017; 179(1): 10-9. doi: 10.1111/bjh.14757.
Schoeman EM, Lopez GH, McGowan EC, Millard GM, O’Brien H, Roulis EV, et al. Evaluation of targeted exome sequencing for 28 protein-based blood group systems, including the homologous gene systems, for blood group genotyping. Transfusion. 2017; 57(4): 1078-88. doi: 10.1111/trf.14054.
Le van Kim C, Mouro I, Chérif-Zahar B, Raynal V, Cherrier C, Cartron JP, et al. Molecular cloning and primary structure of the human blood group RhD polypeptide. Proc Natl Acad Sci U S A. 1992; 89(22): 10925-9. doi: 10.1073/pnas.89.22.10925.
Bonet Bub C, Castilho L. ID CORE XT as a tool formolecular red blood cell typing. Expert Rev Mol Diagn. 2019; 19(9): 777-83. doi: 10.1080/14737159.2019.1656529.
Flesch BK, Scherer V, Just B, Opitz A, Ochmann O,Janson A, et al. Molecular blood group screening in donors from Arabian countries and Iran using high-throughput MALDI-TOF mass spectrometry and PCR-SSP. Transfus Med Hemother. 2020; 47(5): 396-408. doi: 10.1159/000505495.
Hu T, Chitnis N, Monos D, Dinh A. Next-generation sequencing technologies: An overview. Hum Immunol.2021; 82(11): 801-11. doi: 10.1016/j.humimm.2021.02.012.
Quirino MG, Colli CM, Macedo LC, Sell AM, Visentainer JEL. Methods for blood group antigens
detection: cost-effectiveness analysis of phenotyping and genotyping. Hematol Transfus Cell Ther. 2019; 41(1): 44-9. doi: 10.1016/j.htct.2018.06.006.
Ying Y, Zhang J, Hong X, Xu X, He J, Zhu F. The Significance of RHD Genotyping and Characteristic
Analysis in Chinese RhD Variant Individuals. Front Immunol. 2021; 12: 755661. doi: 10.3389/fimmu.2021.755661.
Reid ME, & Lomas-Francis C. The Blood Group Antigens. 2nd Ed. Academic Press; 2010.
Sanger, F., Nicklen, S., & Coulson, A. R. DNA sequencing with chain-terminating inhibitors. Proceedings of the National Academy of Sciences, 1977; 74(12): 5463-7.
Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008; 26(10): 1135-45. doi: 10.1038/nbt1486.
Jain D, Choudhuri J, Chauhan R, Dorwal P, Sharma D, Tiwari AK, Raina V. False negative single antigen bead assay: Is it always an effect of prozone? J Clin Lab Anal. 2018; 32(2): e22237. doi: 10.1002/jcla.22237.
Montemayor-Garcia C, Westhoff CM. The “next generation” reference laboratory? Transfusion. 2018 Feb;58(2):277-279. doi: 10.1111/trf.14483.
Satam H, Joshi K, Mangrolia U, Waghoo S, Zaidi G, Rawool S, Thakare RP, Banday S, Mishra AK, Das G, Malonia SK. Next-Generation Sequencing Technology: Current Trends and Advancements. Biology (Basel). 2023; 12(7): 997. doi: 10.3390/biology12070997.Erratum in: Biology (Basel). 2024; 13(5): 286.
Gleadall NS, Veldhuisen B, Gollub J, Butterworth AS, Ord J, Penkett CJ, et al. Development and validation of a universal blood donor genotyping platform: a multinational prospective study. Blood Adv. 2020; 4(15): 3495-506. doi: 10.1182/bloodadvances.2020001894.
Kwok PY. Methods for genotyping single nucleotide polymorphisms. Annu Rev Genomics Hum Genet. 2001; 2:235-58. doi: 10.1146/annurev.genom.2.1.235.
Syvänen AC. Accessing genetic variation: genotyping single nucleotide polymorphisms. Nat Rev Genet. 2001; 2(12): 930-42. doi: 10.1038/35103535.
Schonewille H, van de Watering LM, Brand A. Additional red blood cell alloantibodies after blood transfusions in a nonhematologic alloimmunized patient cohort: is it time to take precautionary measures? Transfusion. 2006; 46(4): 630-5. doi: 10.1111/ j.1537-2995.2006.00764.x.
Casas J, Friedman DF, Jackson T, Vege S, Westhoff CM, Chou ST. Changing practice: red blood cell typing by molecular methods for patients with sickle cell disease. Transfusion. 2015; 55(6 Pt 2): 1388-93. doi:10.1111/trf.12987.
Chou ST, Flanagan JM, Vege S, Luban NLC, Brown RC, Ware RE, et al. Whole-exome sequencing for RH genotyping and alloimmunization risk in children with sickle cell anemia. Blood Adv. 2017; 1(18): 1414-22. doi: 10.1182/bloodadvances.2017007898.
Chou ST, Evans P, Vege S, Coleman SL, Friedman DF, Keller M, et al. RH genotype matching for transfusion support in sickle cell disease. Blood. 2018; 132(11):1198-207. doi: 10.1182/blood-2018-05-851360.
Chou ST, Alsawas M, Fasano RM, Field JJ, Hendrickson JE, Howard J, et al. American Society of Hematology 2020 guidelines for sickle cell disease: transfusion support. Blood Adv. 2020; 4(2): 327-55. doi: 10.1182/bloodadvances.2019001143.
Lasalle-Williams M, Nuss R, Le T, Cole L, Hassell K, Murphy JR, et al. Extended red blood cell antigen matching for transfusions in sickle cell disease: a review of a 14-year experience from a single center (CME). Transfusion. 2011; 51(8): 1732-9. doi: 10.1111/j. 1537-2995.2010.03045.x.
World Health Organization (WHO). Blood safety and availability. 2023. Available at: https://www.who.int/news-room/fact-sheets/detail/blood-safetyand-availability
Mangmee S, Reamtong O, Kalambaheti T, Roytrakul S, Sonthayanon P. MALDI-TOF mass spectrometry typing for predominant serovars of non-typhoidal Salmonella in a Thai broiler industry. Food Cont. 2020; 113: 107-88. doi.org/10.1016/j.foodcont.2020.107188
Piamsomboon P, Jaresitthikunchai J, Hung TQ, Roytrakul S, Wongtavatchai J. Identification of bacterial pathogens in cultured fish with a custom peptide database constructed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). BMC Vet Res. 2020; 16(1): 52. doi: 10.1186/s12917-020-2274-1.
Sonthayanon P, Jaresitthikunchai J, Mangmee S, Thiangtrongjit T, Wuthiekanun V, Amornchai P, et al. Whole cell matrix assisted laser desorption/ionization time-of-flight mass spectrometry (MALDITOFMS) for identification of Leptospira spp. In Thailand and Lao PDR. PLoS Negl Trop Dis. 2019; 13(4):e0007232. doi: 10.1371/journal.pntd.0007232.
Arpornsuwan T, Buasakul B, Jaresitthikunchai J, Roytrakul S. Potent and rapid antigonococcal activity of the venom peptide BmKn2 and its derivatives against different Maldi biotype of multidrug-resistant Neisseria gonorrhoeae. Peptides. 2014; 53: 315-20. doi: 10.1016/j.peptides.2013.10.020.
Arpornsuwan T, Paveenkittiporn W, Jaresitthikunchai J, Roytrakul S. BAMP-28 antimicrobial peptide against different MALDI biotype of Carbapenam resistant Enterobacteriaceae. Int J Pept Res Ther. 2018; 25(3): 951-60. doi.org/10.1007/s10989-018-9743-4.
Ile R, Mahmoud N. Future Laboratory Medicine: Rapid, Efficient and Affordable Screening for Haemoglobinopathies by MALDI-ToF Mass Spectrometry. Adv Biochem Biotechnol. 2018; ABIO-157. doi: 10.29011/2574-7258. 000057.
Pais RJ, Jardine C, Zmuidinaite R, Lacey J, Butler S, Iles R. Rapid, Affordable and Efficient Screening of Multiple Blood Abnormalities Made Possible Using an Automated Tool for MALDI-ToF Spectrometry Analysis. Appl Sci. 2019; 9(23): 4999. doi.org/10.3390/app9234999.
Iles RK, Iles JK, Abban T, Docherty SM, Nasse M. Method for Detecting Abnormalities in Hemoglobin. WO Patent 2016/030688; July 2019.
Greene DN, Vaughn CP, Crews BO, Agarwal AM. Advances in detection of hemoglobinopathies. ClinChim Acta. 2015; 439: 50-7. doi: 10.1016/j.cca.2014.10.006.
Pedersen M, Vryonidis E, Joensen A, Törnqvist M. Hemoglobin adducts of acrylamide in human blood- What has been done and what is next? Food Chem Toxicol. 2022; 161: 112799. doi: 10.1016/j.fct.2021.112799.
Chen YC, Hsu JF, Chang CW, Li SW, Yang YC, Chao MR, et al. Connecting chemical exposome to human health using high-resolution mass spectrometrybased biomonitoring: Recent advances and future perspectives. Mass Spectrom Rev. 2023; 42(6): 2466-86. doi: 10.1002/mas.21805.
Carlsson H, Rappaport SM, Törnqvist M. Protein Adductomics: Methodologies for Untargeted Screening of Adducts to Serum Albumin and Hemoglobin in Human Blood Samples. High Throughput. 2019; 8(1):6. doi: 10.3390/ht8010006.
Kuleshova ID, Zaripov PI, Poluektov YM, Anashkina AA, Kaluzhny DN, Parshina EY, et al. Changes in Hemoglobin Properties in Complex with Glutathione and after Glutathionylation. Int J Mol Sci. 2023; 24(17): 13557. doi: 10.3390/ijms241713557.
Lockridge O. Overview of Adductomics in Toxicology. Curr Protoc. 2023; 3(2): e672. doi: 10.1002/cpz1.672.
Biroccio A, Urbani A, Massoud R, Di Ilio C, Sacchetta P, Bernardini S, et al. A quantitative method for the analysis of glycated and glutathionylated hemoglobin by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Anal Biochem. 2005; 336(2): 279-88. doi: 10.1016/j.ab.2004.10.002.
Iles RK, Cole LA, Butler SA. Direct analysis of hCGβcf glycosylation in normal and aberrant pregnancy by matrix-assisted laser desorption/ionization time-offlight mass spectrometry. Int J Mol Sci. 2014; 15(6):10067-82. doi: 10.3390/ijms150610067.
Rubino FM, Ottolenghi S, Brizzolari A, Maioli C, Samaja M, Paroni R. Enhanced-Precision Measurement of Glutathionyl Hemoglobin by MALDI-ToF MS. Molecules. 2023; 28(2): 497. doi: 10.3390/molecules28020497.
Rael LT, Bar-Or R, Ambruso DR, Mains CW, Slone DS, Craun ML, et al. The effect of storage on the accumulation of oxidative biomarkers in donated packed red blood cells. J Trauma. 2009; 66(1): 76-81. doi: 10.1097/TA.0b013e318191bfe0.