Assessment of colistin resistance in Enterobacteriaceae: Comparison of the rapid polymyxin NP test, MicroScan system, and broth microdilution in Thailand
Main Article Content
Abstract
Background: Antimicrobial resistance is a global health crisis, with colistin serving as a last-resort antibiotic for the treatment of multidrug-resistant Enterobacteriaceae. However, colistin resistance is becoming increasingly prevalent, necessitating rapid and accurate susceptibility testing.
Objective: This study evaluated the rapid polymyxin NP (RPNP) test and the MicroScan antimicrobial susceptibility system compared to the reference broth microdilution (rBMD) method for detecting colistin resistance in colistin-resistant Enterobacteriaceae (CRE) isolates from clinical setting in Thailand.
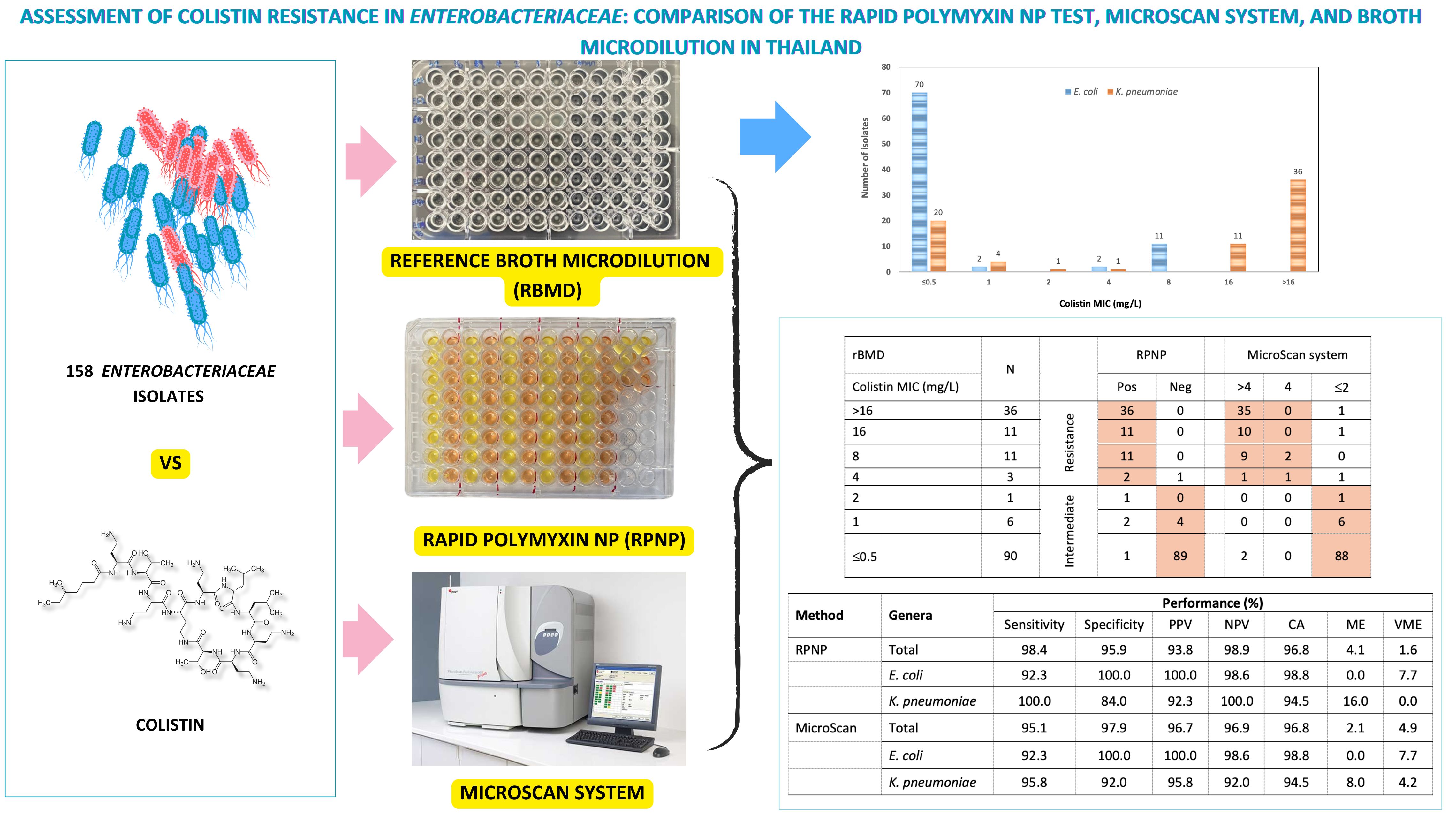
Materials and methods: A total of 158 non-repetitive Enterobacteriaceae isolates (85 Escherichia coli and 73 Klebsiella pneumoniae) were collected from a tertiary hospital in Bangkok between August 2021 and January 2023. Colistin susceptibility was assessed using the RPNP, MicroScan system, and rBMD method following the Clinical and Laboratory Standards Institute (CLSI) guidelines. The categorical agreement (CA), very major errors (VME), and major errors (ME) of colistin resistance test results from the RPNP and MicroScan methods were determined by comparison with the rBMD method.
Results: Among the 158 Enterobacteriaceae isolates, 46.2% were intermediate and 38.6% were colistin-resistant by rBMD. The sensitivity of RPNP was 98.4% and the specificity was 95.9%, compared to 95.1% and 97.9% for MicroScan. Both RPNP and MicroScan achieved high CA (96.8%) with rBMD. Discrepancies, primarily in isolates with borderline MICs (1-4 mg/L), accounted for 4 of the 8 discordant strains. In addition, mobilized colistin resistance-1 (mcr-1) gene were detected in 5 colistin-resistant E. coli isolates with MIC of 8 mg/L.
Conclusion: Both the RPNP and MicroScan systems demonstrated high categorical agreement with the rBMD method, underscoring their reliability as practical tools for routine colistin susceptibility testing in clinical microbiology laboratories where rBMD may not be feasible.
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Personal views expressed by the contributors in their articles are not necessarily those of the Journal of Associated Medical Sciences, Faculty of Associated Medical Sciences, Chiang Mai University.
References
Mancuso G, Midiri A, Gerace E, Biondo C. Bacterial antibiotic resistance: the most critical pathogens. Pathogens. 2021; 10(10): 1310. doi:10.3390/pathogens 10101310
Phumart P, Phodha T, Thamlikitkul V, Riewpaiboon A, Prakongsai P, Limwattananon S. Health and economic impacts of antimicrobial resistant infections in Thailand: a preliminary study. J Health Syst Res. 2012; 6(3): 352-60.
Mondal AH, Khare K, Saxena P, Debnath P, Mukhopadhyay K, Yadav D. A review on colistin resistance: an antibiotic of last resort. Microorganisms. 2024; 12(4): 772. doi: 10.3390/microorganisms12040772
Velkov T, Thompson PE, Nation RL, Li J. Structureactivity relationships of polymyxin antibiotics. J Med Chem. 2010; 53(5): 1898-916. doi:10.1021/jm900999h
Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmidmediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016; 16(2): 161-8. doi:10.1016/S1473-3099(15)00424-7
Dafopoulou K, Zarkotou O, Dimitroulia E, et al. Comparative evaluation of colistin susceptibility testing methods among carbapenem-nonsusceptible Klebsiella pneumoniae and Acinetobacter baumannii clinical isolates. Antimicrob Agents Chemother. 2015; 59(8): 4625-30. doi: 10.1128/AAC.00868-15
Hindler JA, Humphries RM. Colistin MIC variability by method for contemporary clinical isolates of multidrug-resistant Gram-negative bacilli. J Clin Microbiol. 2013; 51(6): 1678-84. doi: 10.1128/JCM.03385-12
Tan TY, Ng SY. Comparison of Etest, Vitek and agar dilution for susceptibility testing of colistin. Clin Microbiol Infect. 2007; 13(5): 541-4. doi: 10.1111/j. 1469-0691.2007.01708.x
Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. 30th Ed. CLSI Supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2020.
Nordmann P, Jayol A, Poirel L. Rapid detection of polymyxin resistance in Enterobacteriaceae. Emerg Infect Dis. 2016; 22(6): 1038-43. doi: 10.3201/eid2206. 151840
Mitton B, Kingsburgh C, Kock MM, Mbelle NM, Strydom K. Evaluation of an in-house colistin NP test for use in resource-limited settings. J Clin Microbiol. 2019; 57(10): e00501-19. doi: 10.1128/JCM.00501-19
Simar S, Sibley D, Ashcraft D, Pankey G. Evaluation of the rapid Polymyxin NP test for polymyxin B resistance detection using Enterobacter cloacae and Enterobacter aerogenes isolates. J Clin Microbiol. 2017; 55(10): 3016-20. doi: 10.1128/JCM.00934-17
Jayol A, Kieffer N, Poirel L, et al. Evaluation of the Rapid Polymyxin NP test and its industrial version for the detection of polymyxin-resistant Enterobacteriaceae. Diagn Microbiol Infect Dis. 2018; 92(2): 90-4. doi: 10.1016/j.diagmicrobio.2018.05.006
Yainoy S, Hiranphan M, Phuadraksa T, Eiamphungporn W, Tiengrim S, Thamlikitkul V. Evaluation of the rapid polymyxin NP test for detection of colistin susceptibility in Enterobacteriaceae isolated from Thai patients. Diagn Microbiol Infect Dis. 2018; 92(2): 102-6. doi: 10.1016/j.diagmicrobio.2018.05.009
Pfennigwerth N, Kaminski A, Korte-Berwanger M, et al. Evaluation of six commercial products for colistin susceptibility testing in Enterobacterales. Clin Microbiol Infect. 2019; 25(11): 1385-9. doi: 10.1016/j.cmi.2019. 03.017
Jayol A, Nordmann P, André C, Poirel L, Dubois V. Evaluation of three broth microdilution systems to determine colistin susceptibility of gram-negative bacilli. J Antimicrob Chemother. 2018; 73(5): 1272-8. doi: 10.1093/jac/dky012
Clinical and Laboratory Standards Institute. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 10th Ed. CLSI document M07-A10. Wayne, PA: Clinical and Laboratory Standards Institute; 2015.
The European Committee on Antimicrobial Susceptibility Testing. Routine and extended internal quality control for MIC determination and disk diffusion as recommended by EUCAST. Version 15.0, 2025. Available from: http:// www.eucast.org
Clinical and Laboratory Standards Institute. Verification of commercial microbial identification and antimicrobial susceptibility testing systems: M52. 1st Ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2015.
Conceição-Neto OC, da Costa BS, Pontes LS, Santos ICO, Silveira MC, Cordeiro-Moura JR, et al. Difficulty in detecting low levels of polymyxin resistance in clinical Klebsiella pneumoniae isolates: evaluation of Rapid Polymyxin NP test, Colispot Test and SuperPolymyxin medium. New Microbes New Infect. 2020; 36: 100722. doi: 10.1016/j.nmni.2020.100722
Eiamphungporn W, Yainoy S, Jumderm C, et al. Prevalence of the colistin resistance gene mcr-1 in colistin- resistant Escherichia coli and Klebsiella pneumoniae isolated from humans in Thailand. J Glob Antimicrob Resist. 2018; 15: 32-5. doi: 10.1016/j.jgar. 2018.06.007
Poirel L, Jayol A, Nordmann P. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. 2017; 30(2): 557-96. doi:10.1128/CMR.00064-16
Jayol A, Nordmann P, Lehours P, Poirel L, Dubois V. Comparison of methods for detection of plasmidmediated and chromosomally encoded colistin resistance in Enterobacteriaceae. Clin Microbiol Infect. 2018; 24(2): 175-9. doi: 10.1016/j.cmi.2017.06.002
Quan J, Li X, Chen Y, et al. Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect Dis. 2017; 17(4): 400-10. doi: 10.1016/S1473-3099(16)30528-X
Zhang H, Zhao D, Quan J, Hua X, Yu Y. mcr-1 facilitated selection of high-level colistin-resistant mutants in Escherichia coli. Clin Microbiol Infect. 2019; 25(4): 517.e1-517.e4. doi: 10.1016/j.cmi.2018.12.014
Bastidas-Caldes C, de Waard JH, Salgado MS, Villacís MJ, Coral-Almeida M, Yamamoto Y, et al. Worldwide prevalence of mcr-mediated colistin-resistant Escherichia coli in isolates of clinical samples, healthy humans, and livestock: a systematic review and meta-analysis. Pathogens. 2022; 11(6): 659. doi: 10.3390/pathogens 11060659
Ling Z, Yin W, Shen Z, Wang Y, Shen J, Walsh TR. Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. J Antimicrob Chemother. 2020; 75(11): 3087-95. doi: 10.1093/jac/dkaa205