Detection and characterization of SARS-CoV-2 variants in Thailand from 2020-2023 using next-generation sequencing
Main Article Content
Abstract
Background: The evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has led to multiple variants with significant public health implications.
Objective: This study aimed to detect and characterize SARS-CoV-2 genomic variants in Thailand using next-generation sequencing (NGS) from 2020 to 2023.
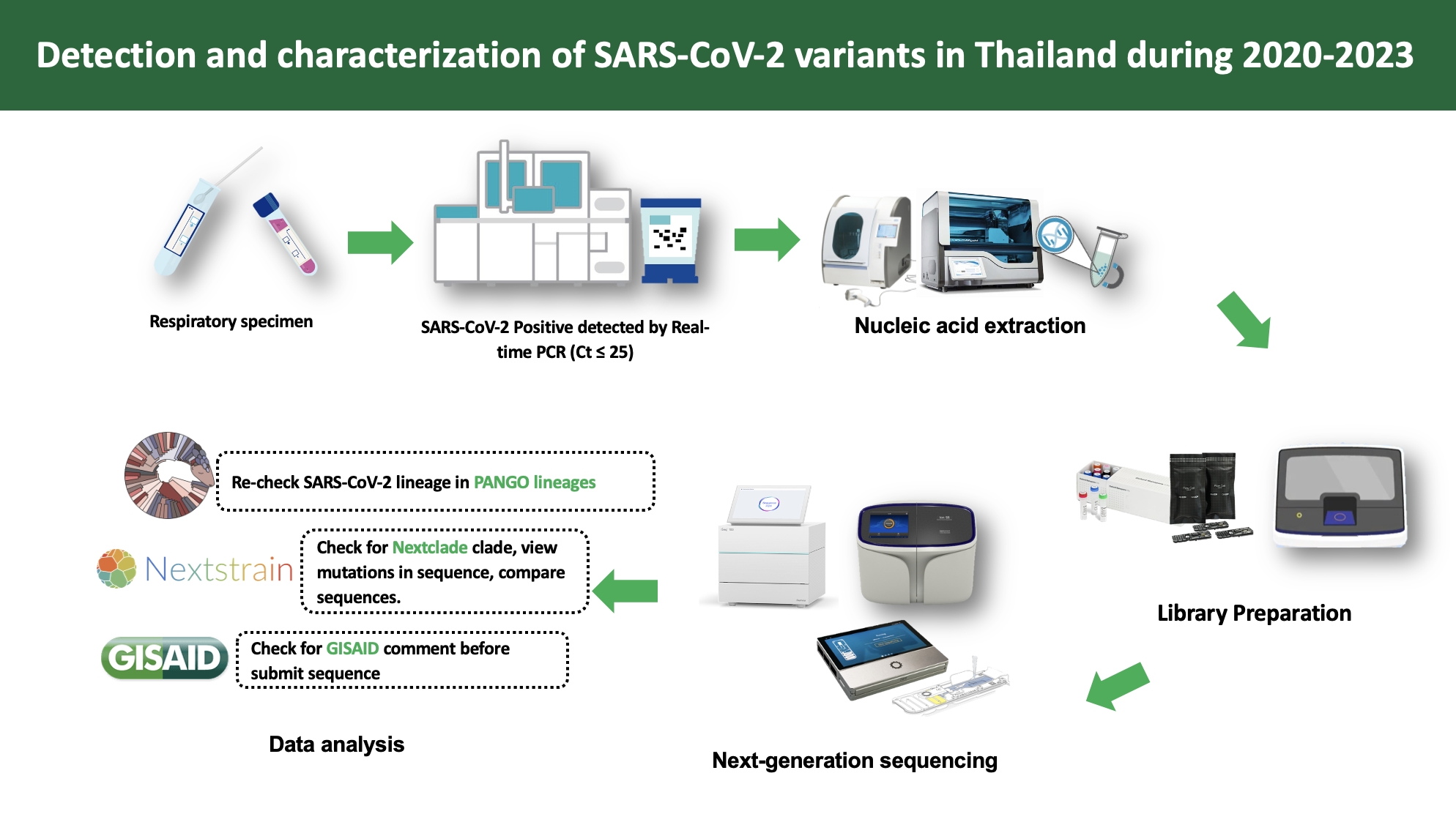
Materials and methods: A total of 2,592 oro/nasopharyngeal swab specimens with high viral loads (cycle threshold; Ct≤25.0) were collected from SARS-CoV2-positive patients in Nonthaburi, Nakhon Phanom, and Tak Provinces. NGS was performed using Ion Torrent, Oxford Nanopore Technologies, and Illumina platforms to identify and analyze SARS-CoV-2 variants.
Results: The sequencing revealed various SARS-CoV-2 variants circulating in Thailand, with 2,592 lineages identified. The Omicron variant was predominant, accounting for 93.44% (2,422/2,592). The most common Omicron subvariants were BA.2 (54.86%, 1,422/2,592), BA.5 (20.22%, 524/2,592), and XBB (14.51%, 376/2,592). The Delta variant constituted 2.66% (69/2,592) of the cases, with AY.30 (1.20%, 31/2,592) and AY.85 (1.27%, 33/2,592) being the most frequent sub-lineages. The Alpha variant (B.1.1.7) was detected in 33 cases (1.27%), and the Beta variant (B.1.351) in 1 case (0.04%).
Conclusion: The findings reveal a significant shift in the SARS-CoV-2 variant landscape in Thailand, with Omicron becoming the dominant strain post-2022. Continuous genomic surveillance is critical for tracking viral evolution, informing public health strategies, and supporting pandemic control efforts in Thailand and Southeast Asia.
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Personal views expressed by the contributors in their articles are not necessarily those of the Journal of Associated Medical Sciences, Faculty of Associated Medical Sciences, Chiang Mai University.
References
Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, et al. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020; 182(4): 812-27.e19. doi: 10.1016/j.cell.2020.06.043.
Callaway E. The coronavirus is mutating - does it matter?. Nature. 2020; 585(7824): 174-7. doi: 10.1038/d41586- 020-02544-6.
Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet. 2020; 395(10224): 565-74. doi: 10.1016/S0140-6736(20)30251-8.
Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, et al. Nextstrain: real-time tracking of pathogen evolution. Kelso J, editor. Bioinformatics. 2018; 34(23): 4121-3. doi: 10.1093/bioinformatics/ bty407.
Paden CR, Tao Y, Queen K, Zhang J, Li Y, Uehara A, et al. Rapid, sensitive, full-genome sequencing of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg Infect Dis. 2020; 26(10): 2401-5. doi: 10.3201/ eid2610.201800.
World Health Organization. SARS-CoV-2 genomic sequencing for public health goals: interim guidance, 8 January 2021 [Internet]. Geneva: World Health Organization; 2021 [cited 2024 Dec 11]. Available from: https://iris.who.int/bitstream/handle/10665/ 338483/WHO-2019-nCoV-genomic_sequencing2021.1-eng.pdf?sequence=1&isAllowed=y.
Okada P, Buathong R, Phuygun S, Thanadachakul T, Parnmen S, Wongboot W, et al. Early transmission patterns of coronavirus disease 2019 (COVID-19) in travellers from Wuhan to Thailand, January 2020. Eurosurveillance. 2020;25(8). doi: 10.2807/1560- 7917.ES.2020.25.8.2000097.
Department of Disease Control, Ministry of Public Health, Thailand. First confirmed case of COVID-19 in Thailand. 2020.
World Health Organization. WHO Coronavirus (COVID-19) Dashboard: Thailand [Internet]. Geneva: World Health Organization; [cited 2024 Nov 29]. Available from: https://covid19.who.int/region/ searo/country/th.
Sirijatuphat R, Suputtamongkol Y, Angkasekwinai N, Horthongkham N, Chayakulkeeree M, et al. Epidemiology, clinical characteristics, and treatment outcomes of patients with COVID-19 at Thailand’s university-based referral hospital. BMC Infect Dis. 2021; 21(1): 382. doi: 10.1186/s12879-021-06081-z.
World Health Organization. Thailand COVID-19 case study, 20 September [Internet]. [cited 2024 Dec 11]. Available from: C:/Users/hugo2/Downloads/ thailand-c19-case-study-20-september.pdf.
Sooksawasdi Na Ayudhya S, Wongsanuphat S, Surareungchai W, Hirankarn N. Emergence and spread of SARS-CoV-2 variants in Thailand: A genomic surveillance study. Sci Rep. 2024; 14: 56646. doi. org/10.1038/s41598-024-56646-6.
Bunyong R, Chantarangsu S, Sudsai T, Suwannakarn K, Sripramote T, et al. Genomic characterization of SARS-CoV-2 variants in Thailand and implications for vaccine efficacy. Viruses. 2023; 15(6): 1394. doi. org/10.3390/v15061394.
Tongsima S, Surapolchai P, Sookrung N, Sakornkarn K, Poovorawan Y. Regional dynamics and the impact of cross-border movements on SARS-CoV-2 spread in Southeast Asia. Microb Genom. 2023; 9(5): 001170. doi.org/10.1099/mgen.0.001170.
Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016; 17(6): 333-51. doi: 10.1038/nrg.2016.49.
Reuter JA, Spacek DV, Snyder MP. High-throughput sequencing technologies. Mol Cell. 2015; 58(4): 586- 97. doi: 10.1016/j.molcel.2015.05.004.
Thermo Fisher Scientific. SuperScript™ IV One-Step RT-PCR System [Internet]. [cited 2024 Nov 29]. Available from: https://www.thermofisher.com/order/ catalog/product/A49670.
Thermo Fisher Scientific. Qubit™ 4 Fluorometer [Internet]. [cited 2024 Nov 29]. Available from: https://www.thermofisher.com/order/catalog/product/Q33216.
Thermo Fisher Scientific. Ion AmpliSeq™ SARS-CoV-2 Insight Research Assay for Genexus™ System User Guide [Internet]. 2020 [cited 2024 Nov 29]. Available from: https://assets.thermofisher.com/TFS-Assets/ LSG/manuals/MAN0024915_IAInsight_Cov2forGS_ UG.pdf.
Oxford Nanopore Technologies. Ligation Sequencing Kit SQK-LSK109 [Internet]. [cited 2024 Nov 29]. Available from: https://store.nanoporetech.com/ligation-sequencingkit-sqk-lsk109.html
Illumina. COVIDSeq Data Sheet [Internet]. 2020 [cited 2024 Nov 29]. Available from: https://www.illumina. com/content/dam/illumina/gcs/assembled-assets/ marketing-literature/covidseq-data-sheet-m-gl-00243/ illumina-covidseq-data-sheet-m-gl-00243.pdf.
Illumina. Respiratory Virus Oligo Panel [Internet]. [cited 2024 Nov 29]. Available from: https://www. illumina.com/products/by-type/sequencing-kits/ library-prep-kits/respiratory-virus-oligo-panel.html.
Szargut M, Cytacka S, Serwin K, Urbańska A, Gastineau R, Parczewski M, Ossowski A. SARS-CoV-2 wholegenome sequencing by Ion S5 Technology-Challenges, protocol optimization and success rates for different strains. Viruses. 2022; 14(6): 1230. doi: 10.3390/v140 61230.
Aksamentov I, Roemer C, Hodcroft EB, Neher RA. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J Open Source Softw. 2021; 6: 3773. doi.org/10.21105/joss.03773.
Sample Size Calculator. Confidence interval for a proportion [Internet]. [cited 2024 Nov 29]. Available from: https://sample-size.net/confidence-intervalproportion/.
Tongsima S, Sooksawasdi Na Ayudhya S, Bunyong R. Whole-genome sequencing of SARS-CoV-2: Implications for variant detection and surveillance. J Infect Dis. 2023; 227: 698-707. doi: 10.1016/j.ijid. 2022.05.041..
Ashford A, Goldsmith CS, Perkins MD, et al. Genomic characterization and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020; 395: 565-74.
Worobey M, Pekar J, Larsen BB, et al. The emergence and early spread of SARS-CoV-2 in Europe and North America. Science. 2020; 370: 564-70. doi: 10.1126/ science.abc8169.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018; 35: 1547-9. doi: 10.1093/molbev/msy096.
Puenpa J, Rattanakomol P, Saengdao N, Chansaenroj J, Yorsaeng R, et al. Molecular characterisation and tracking of severe acute respiratory syndrome coronavirus 2 in Thailand, 2020-2022. Arch Virol. 2023; 168(1): 26. doi: 10.1007/s00705-022-05666-6.
Wang Y, Wong NS, Lee SS, Ip DK, Wu P, Lau EHY, et al. Clinical profile analysis of SARS-CoV-2 community infections during periods with omicron BA.2, BA.4/5, and XBB dominance in Hong Kong: a prospective cohort study. Lancet Infect Dis. 2024: S1473-3099 (24000574. doi:10.1016/S1473-3099(24) 00574-7.