Prevalence and antimicrobial resistance of Salmonella isolated from pork in the Northern part of Thailand
Main Article Content
Abstract
Background: Salmonella is one of the most common foodborne bacteria frequently isolated from pork. It pose significant public health risks due to its potential to harbor antimicrobial resistance (AMR) genes. The emergence and spread of multidrug-resistant (MDR) Salmonella strains are of particular concern as these can be transferred to humans through food or contaminated environments.
Objective: To determine Salmonella’s prevalence and antimicrobial resistance determinants from pork at retail markets in the Northern part of Thailand.
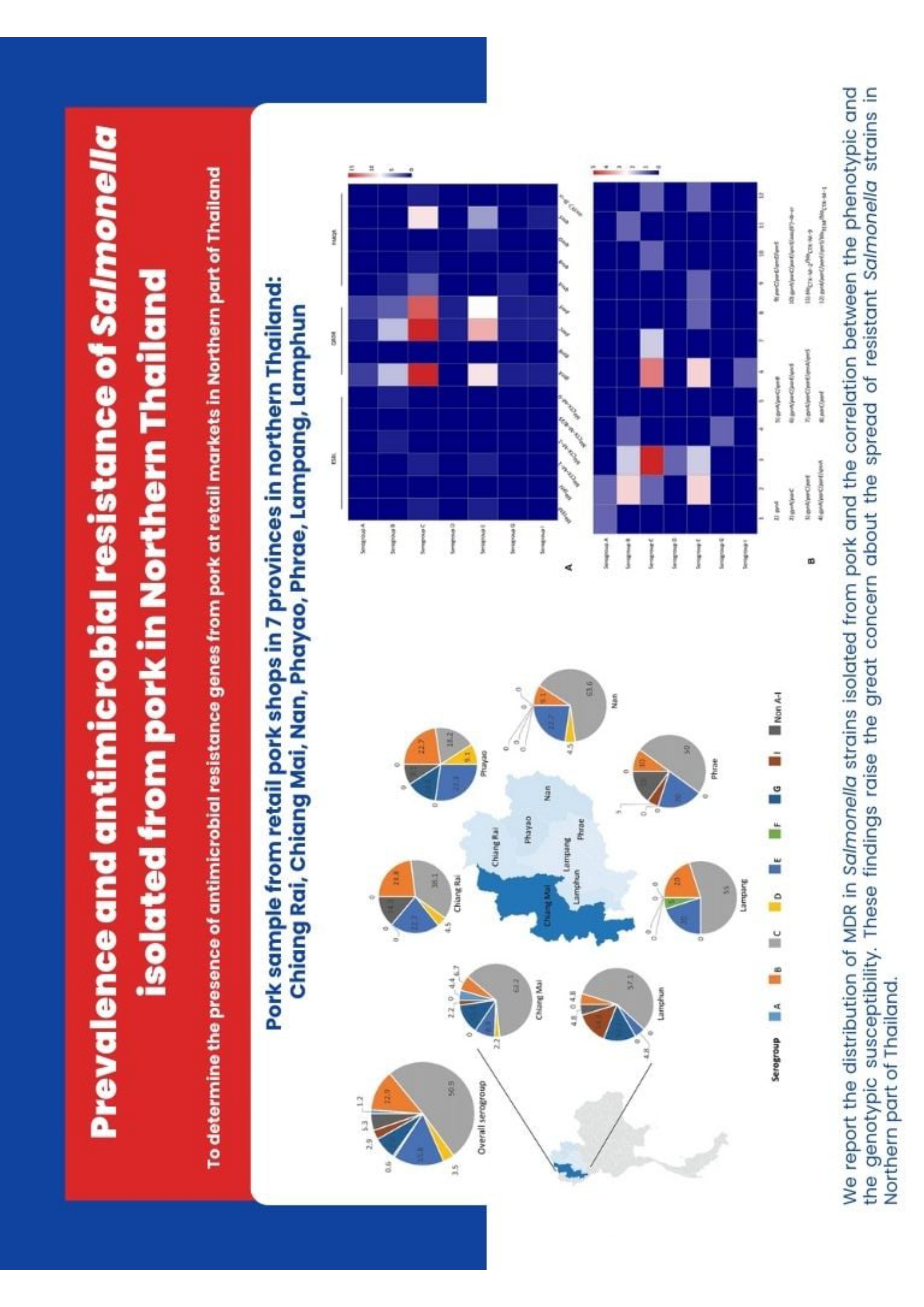
Materials and methods: Salmonella was isolated and identified from 173 pork samples. The isolates were serotyped using the slide agglutination test with somatic- (O) and flagellar- (H) antigens, and tested for antimicrobial susceptibility using 23 different antimicrobials using the disc diffusion method. By polymerase chain reaction (PCR), the extended-spectrum β-lactamases (ESBL) genes (blaTEM, blaSHV, blaCTX-M-1, blaCTX-M-2, blaCTX-M-8/25 and blaCTX-M-9), quinolone-resistance determining region (QRDR) including gyrA, gyrB, parC and parE, plasmid-mediated quinolone resistance (PMQR) determinants including qnrA, qnrB, qnrD, qnrS, and aac(6’)-Ib-cr were determined.
Results: In this study, 98.8% of the samples were identified as Salmonella. Among these, 72.5% of isolates showed resistance to at least 1 antimicrobial agent and 48.5% were MDR. Ampicillin/tetracycline/trimethoprim-sulfamethoxazole (AMP/ TE/SXT) was the most common phenotypic resistance pattern. The results revealed that 4 ESBL genes (blaTEM, blaCTX-M-1, blaCTX-M-2, and blaCTX-M-9) were detected in 3 isolates- identified as ESBL producers. Moreover, all 9 quinolone-mediated resistance determinants were observed in quinolone resistance isolates.
Conclusion: The results demonstrated that most MDR isolates harbored quinolone resistance determinants. We report the distribution of MDR in Salmonella strains isolated from pork and the correlation between the phenotypic and the genotypic susceptibility. These findings raise great concern about the spread of resistant Salmonella strains in the Northern part of Thailand.
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Personal views expressed by the contributors in their articles are not necessarily those of the Journal of Associated Medical Sciences, Faculty of Associated Medical Sciences, Chiang Mai University.
References
Dewey-Mattia D MK, Hall AJ, Wise ME, Crowe SJ. Surveillance for Foodborne Disease Outbreaks — United States, 2009-2015. MMWR Surveill Summ. 2018; 67(10): 1-11. doi: 10.15585/ mmwr.ss6710a1
Aslam B, Khurshid M, Arshad MI, Muzammil S, Rasool M, Yasmeen N, et al. Antibiotic resistance: One health one world outlook. Front Cell Infect Microbiol. 2021; 11: 771510. doi: 10.3389/ fcimb.2021.771510.
Hosain MZ, Kabir SML, Kamal MM. Antimicrobial uses for livestock production in developing countries. Vet World. 2021; 14(1): 210-21. doi: 10.14202/vetworld. 2021.210-221.
Lay KK, Jeamsripong S, Sunn KP, Angkititrakul S, Prathan R, Srisanga S, et al. Colistin resistance and ESBL production in Salmonella and Escherichia coli from pigs and pork in Thailand, Cambodia, Lao PDR, and Myanmar border area Antibiotics (Basel). 2021; 10(6): 657. doi: 10.3390/antibiotics 10060657.
Pungpian C, Lee S, Trongjit S, Sinwat N, Angkititrakul S, Prathan R, et al. Colistin resistance and plasmidmediated mcr genes in Escherichia coli and Salmonella isolated from pigs, pig carcass and pork in Thailand, Lao PDR and Cambodia border provinces. J Vet Sci. 2021; 22(5): e68. doi: 10.4142/jvs.2021.22.e68.
Meunsene D, Eiamsam-Ang T, Patchanee P, Pascoe B, Tadee P, Tadee P. Molecular evidence for cross boundary spread of Salmonella spp. in meat sold at retail markets in the middle Mekong basin area. PeerJ. 2021; 9: e11255. doi: 10.7717/peerj.11255.
Niyomdecha N, Mungkornkaew N, Samosornsuk W. Serotypes and antimicrobial resistance of Salmonella Enterica isolated from pork, chicken meat and lettuce, Bangkok and Central Thailand. Southeast Asian J Trop Med Public Health. 2016; 47(1): 31-9.
Prasertsee T, Chokesajjawatee N, Santiyanont P, Chuammitri P, Deeudom M, Tadee P, et al. Quantification and rep-PCR characterization of Salmonella spp. in retail meats and hospital patients in Northern Thailand. Zoonoses Public Health. 2019; 66(3): 301-9. doi: 10.1111/ zph.12565.
Bergspica I, Kaprou G, Alexa EA, Prieto M, AlvarezOrdonez A. Extended spectrum beta-lactamase (ESBL) producing Escherichia coli in pigs and pork meat in the European Union. Antibiotics (Basel). 2020; 9(10): 678. doi: 10.3390/antibiotics9100678.
Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis. 2006; 6(10): 629-40. doi: 10.1016/ S1473-3099(06)70599-0.
Wiener ES, Heil EL, Hynicka LM, Johnson JK. Are fluoroquinolones appropriate for the treatment of extended spectrum beta-lactamase-producing gram-negative bacilli? J Pharm Technol. 2016; 32(1): 16-21. doi: 10.1177/8755122515599407.
World health organization. Critically important antimicrobials for human medicine: WHO: Geneva, Switzerland; 2012 Available from: http://apps.who. int/iris/bitstream/10665/77376/1/9789241504 485_eng.pdf? ua=1&ua=1.
Phongaran D, Khang-Air S, Angkititrakul S. Molecular epidemiology and antimicrobial resistance of Salmonella isolates from broilers and pigs in Thailand. Vet World. 2019; 12(8): 1311-8. doi: 10.14202/vetworld. 2019.1311-1318.
Nuanmuang N, Kummasook A. Prevalence and antimicrobial resistance of Salmonella in minced pork from retail shops around the University of Phayao, Thailand. Asian Health, Science and Technology Reports (AHSTR). 2018; 26(4): 9-16. doi: 10.14456/ nujst.2018.17.
International organization for standardization. ISO 6579-1:1993. Microbiology of food and animal feeding stuffs Horizontal method for the detection of Salmonella spp. Part 1: General guidelines. Geneva: ISO; 1993.
World Health Organization collaborating centre for reference and research on Salmonella. Antigenic formulae of the Salmonella serovars. 2007. Available from: https://www.pasteur.fr/sites/default/files/veng_0. pdf.
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012; 18(3): 268-81. doi: 10.1111/ j.1469-0691.2011.03570.x.
Wang J, Li Y, Xu X, Liang B, Wu F, Yang X, et al. Antimicrobial resistance of Salmonella enterica Serovar Typhimurium in Shanghai, China. Front Microbiol. 2017; 8: 510. doi: 10.3389/ fmicb.2017. 00510.
Kong-Ngoen T, Santajit S, Tunyong W, Pumirat P, Sookrung N, Chaicumpa W, et al. Antimicrobial resistance and virulence of non-Typhoidal Salmonella from retail foods marketed in Bangkok, Thailand. Foods. 2022; 11(5): 661. doi: 10.3390/foods11050661.
Rortana C, Nguyen-Viet H, Tum S, Unger F, Boqvist S, Dang-Xuan S, et al. Prevalence of Salmonella spp. and Staphylococcus aureus in chicken meat and pork from Cambodian markets. Pathogens. 2021; 10(5): 556. doi: 10.3390/pathogens10050556.
Nguyen DT, Kanki M, Nguyen PD, Le HT, Ngo PT, Tran DN, et al. Prevalence, antibiotic resistance, and extended-spectrum and AmpC beta-lactamase productivity of Salmonella isolates from raw meat and seafood samples in Ho Chi Minh City, Vietnam. Int J Food Microbiol. 2016; 236: 115-22. doi: 10.1016/ j.ijfoodmicro.2016.07.017.
Dewey-Mattia D, Manikonda K, Hall AJ, Wise ME, Crowe SJ. Surveillance for foodborne disease outbreaks - United States, 2009-2015. MMWR Surveill Summ.2018; 67(10): 1-11. doi: 10.15585/mmwr.ss6710a1.
Zhang L, Fu Y, Xiong Z, Ma Y, Wei Y, Qu X, et al. Highly prevalent multidrug-resistant Salmonella from chicken and pork meat at retail markets in Guangdong, China. Front Microbiol. 2018; 9: 2104. doi: 10.3389/fmicb. 2018.02104.
Brenner FW, Villar RG, Angulo FJ, Tauxe R, Swaminathan B. Salmonella nomenclature. J Clin Microbiol. 2000; 38(7): 2465-7. doi: 10.1128/JCM.38.7.2465-2467.2000.
Fuche FJ, Sow O, Simon R, Tennant SM. Salmonella Serogroup C: Current status of vaccines and why they are needed. Clin Vaccine Immunol. 2016; 23(9): 737- 45. doi: 10.1128/cvi.00243-16.
Trongjit S, Angkititrakul S, Tuttle RE, Poungseree J, Padungtod P, Chuanchuen R. Prevalence and antimicrobial resistance in Salmonella enterica isolated from broiler chickens, pigs and meat products in Thailand-Cambodia border provinces. Microbiol Immunol. 2017; 61(1): 23-33. doi: 10.1111/1348-04 21.12462.
Pham TDM, Ziora ZM, Blaskovich MAT. Quinolone antibiotics. Medchemcomm. 2019; 10(10): 1719-39. doi: 10.1039/c9md00120d.
Mercer MA. Quinolones, Including Fluoroquinolones, Use in Animals MSD Veterinary Manual: MSD Veterinary Manual; 2022. Available from: https://www.msdvetmanual.com/pharmacology/antibacterial-agents/ quinolones-including-fluoroquinolones-use-in-animals.
Yin X, Dudley EG, Pinto CN, M’Ikanatha N M. Fluoroquinolone sales in food animals and quinolone resistance in non-typhoidal Salmonella from retail meats: United States, 2009-2018. J Glob Antimicrob Resist. 2022; 29: 163-7. doi: 10.1016/j.jgar.2022.03.005.
Cameron-Veas K, Fraile L, Napp S, Garrido V, Grillo MJ, Migura-Garcia L. Multidrug resistant Salmonella enterica isolated from conventional pig farms using antimicrobial agents in preventative medicine programmes. Vet J. 2018; 234: 36-42. doi: 10.1016/j. tvjl.2018.02.002.
Hengkrawit K, Tangjade C. Prevalence and trends in antimicrobial susceptibility patterns of multi-drugresistance non-Typhoidal Salmonella in Central Thailand, 2012-2019. Infect Drug Resist. 2022; 15: 1305-15. doi: 10.2147/IDR.S355213.
Chen Y, Liu L, Guo Y, Chu J, Wang B, Sui Y, et al. Distribution and genetic characterization of fluoroquinolone resistance gene qnr among Salmonella strains from chicken in China. Microbiol Spectr. 2024; 12(4): e0300023. doi: 10.1128/spectrum. 03000-23.
Nwosu UO, Ibiam FA, Amadi-Ibiam CO, Iroha CS, Edemekong CI, Peter IU, et al. Fecal carriage of extended spectrum beta-lactamase and fluoroquinolone resistant gene in non-typhoidal Salmonella enterica isolates from food-producing animals and humans. Journal of Drug Delivery and Therapeutics. 2023; 13(9): 128-34. doi: 10.22270/jddt.v13i9.5964.
Kuang D, Zhang J, Xu X, Shi W, Chen S, Yang X, et al. Emerging high-level ciprofloxacin resistance and molecular basis of resistance in Salmonella enterica from humans, food and animals. Int J Food Microbiol. 2018; 280: 1-9. doi: 10.1016/j.ijfoodmicro.2018.05.001.
Qian H, Cheng S, Liu G, Tan Z, Dong C, Bao J, et al. Discovery of seven novel mutations of gyrB, parC and parE in Salmonella Typhi and Paratyphi strains from Jiangsu Province of China. Sci Rep. 2020; 10(1): 7359. doi: 10.1038/s41598-020-64346-0.
Ngoi ST, Thong KL. High resolution melting analysis for rapid mutation screening in gyrase and Topoisomerase IV genes in quinolone-resistant Salmonella enterica. Biomed Res Int. 2014; 2014: 718084. doi: 10.1155/2014/718084.
Gu Y, Huang L, Wu C, Huang J, Hao H, Yuan Z, et al. The evolution of fluoroquinolone resistance in Salmonella under exposure to sub-inhibitory concentration of enrofloxacin. Int J Mol Sci. 2021; 22(22): 12218. doi: 10.3390/ijms222212218.
Adel WA, Ahmed AM, Hegazy Y, Torky HA, Shimamoto T. High prevalence of ESBL and plasmid-mediated quinolone resistance genes in Salmonella enterica isolated from retail meats and slaughterhouses in Egypt. Antibiotics (Basel). 2021; 10(7) : 881. doi: 10.3390/ antibiotics10070881.
Eiamsam-ang T, Tadee P, Pascoe B, Patchanee P. Genome-based analysis of infrequent Salmonella serotypes through the Thai pork production chain. Frontiers in Microbiology. 2022; 13: 968695. doi: 10.3389/fmicb.2022.968695