A comparative study of pre-processing methods to improve glioma segmentation performance in brain MRI using deep learning
Main Article Content
Abstract
Background: Glioma is the most common brain tumor in adult patients and requires accurate treatment. The delineation of tumor boundaries must be accurate and precise, which is crucial for treatment planning. Currently, delineating boundaries for tumors is a tedious, time-consuming task and may be prone to human error among oncologists. Therefore, artificial intelligence plays a vital role in reducing these problems.
Objective: This study aims to find a relationship between improving image enhancement and evaluating the performance of deep learning models for segmenting glioma image data on brain MRI images.
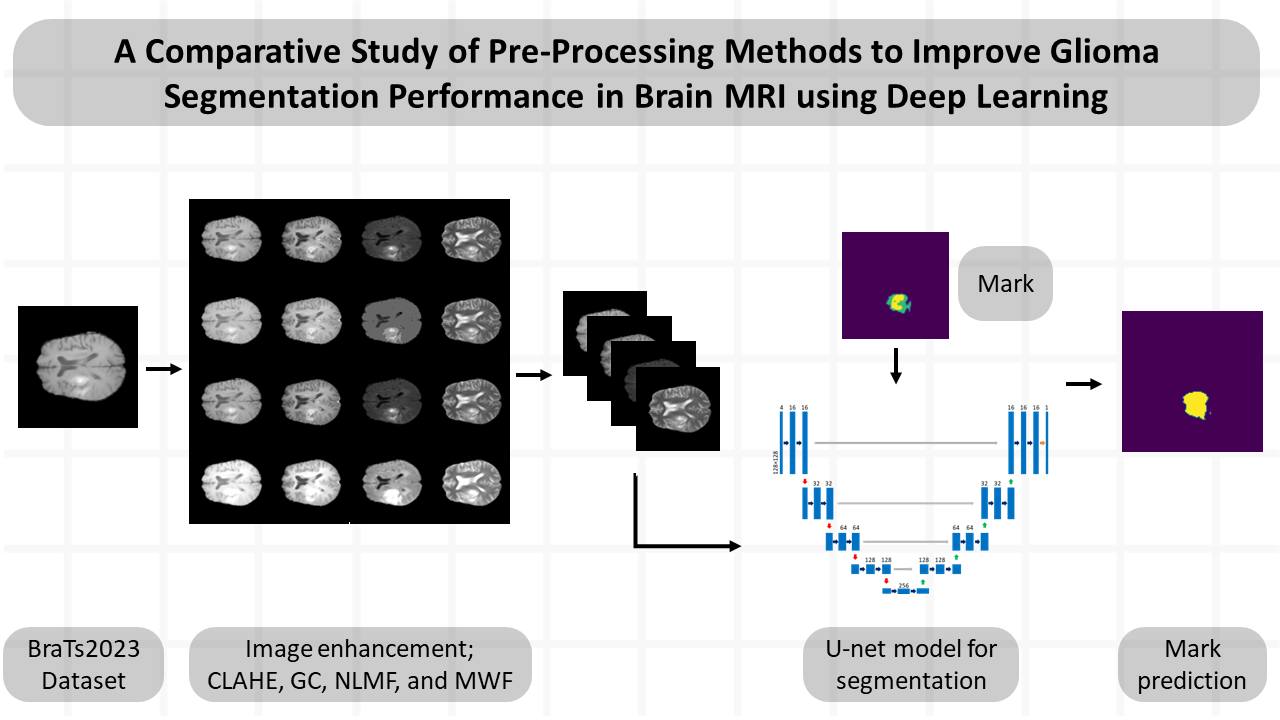
Materials and methods: The BraTs2023 dataset was used in this study. The image dataset was converted from three dimensions to two dimensions and then subjected to pre-processing via four image enhancement techniques, including contrast-limited adaptive histogram equalization (CLAHE), gamma correction (GC), non-local mean filter (NLMF), and median and Wiener filter (MWF). Subsequently, it was evaluated for structural similarity index (SSIM) and mean squared error. The deep learning segmentation model was created using the U-Net architecture and assessed for dice similarity coefficient (DSC), accuracy, precision, recall, F1-score, and Jaccard index for tumor segmentation.
Results: The performance of enhanced image results for CLAHE, GC, NLMF, and MWF techniques shows SSIM values of 0.912, 0.905, 0.999, and 0.911, respectively. The dice similarity coefficient (DSC) for segmentation without image enhancement was 0.817. The DSC of segmentation for CLAHE, GC, NLMF, and MWF techniques were 0.818, 0.812, 0.820, and 0.797, respectively.
Conclusion: The enhanced image technique could affect the performance of tumor segmentation. by the enhanced image for use in a trained model may increase or decrease performance depending on the chosen image enhancement technique and the parameters determined by each method.
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Personal views expressed by the contributors in their articles are not necessarily those of the Journal of Associated Medical Sciences, Faculty of Associated Medical Sciences, Chiang Mai University.
References
Baid U, Ghodasara S, Mohan S, Bilello M, Calabrese E, Colak E, et al. The rsna-asnr-miccai brats 2021 benchmark on brain tumor segmentation and radiogenomic classification. arXiv. 2021; 2107.02314. doi.org/10.48550/arXiv.2107.02314
Lin M, Momin S, Lei Y, Wang H, Curran WJ, Liu T, et al. Fully automated brain tumor segmentation from multiparametric MRI using 3D context deep supervised U-Net. Med Phys. 2021; 48(8): 4365-74. doi.org/10.1002/mp.15032.
Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imaging. 2014; 34(10): 1993-2024. doi: 10.1109/TMI.2014.2377694.
Lin M, Momin S, Lei Y, Wang H, Curran WJ, Liu T, et al. Fully automated brain tumor segmentation from multiparametric MRI using 3D context deep supervised U-Net. Med Phys. 2021; 48(8): 4365-74. doi.org/10.1002/mp.15032.
Knuth F, Adde IA, Huynh BN, Groendahl AR, Winter RM, Negård A, et al. MRI-based automatic segmentation of rectal cancer using 2D U-Net on two independent cohorts. Acta Oncol. 2022; 61(2): 255-63. doi.org/10.1080/0284186X. 2021.2013530.
Zhao C, Zhao Z, Zeng Q, Feng Y. MVP U-Net: Multiview pointwise U-Net for brain tumor segmentation. InBrainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: BrainLes 2020, Lecture Notes in Computer Science, October 4, 2020: Lima, Peru, 12659: 93-103. Springer, Cham. doi.org/10.1007/978-3-030-72087-2_9
Güneş AM, van Rooij W, Gulshad S, Slotman B, Dahele M, Verbakel W. Impact of imperfection in medical imaging data on deep learning-based segmentation performance: An experimental study using synthesized data. Med Phys. 2023; 50(10): 6421-32. doi: 10.1002/mp.16437
Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby JS, et al. Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data. 2017; 4(1): 1-3. doi.org/10.1038/sdata.2017.117.
Wen H, Qi W, Shuang L. Medical X-ray image enhancement based on wavelet domain homomorphic filtering and CLAHE. Proceedings of In 2016 International Conference on Robots & Intelligent System (ICRIS), IEEE. 2016 Aug 27: 249-54. doi: 10.1109/ICRIS.2016.50.
Zuiderveld K. Contrast limited adaptive histogram equalization. Graphics gems. 1994; 474-85. doi.org/10.1016/B978-0-12-336156-1.50061-6.
Cao G, Huang L, Tian H, Huang X, Wang Y, Zhi R. Contrast enhancement of brightness-distorted images by improved adaptive gamma correction. Comput Electr Eng. 2018; 66: 569-82. doi.org/10.1016/j.compeleceng.2017.09.012.
Huang Z, Zhang T, Li Q, Fang H. Adaptive gamma correction based on cumulative histogram for enhancing near-infrared images. Infrared Phys Technol. 2016; 79: 205-15. doi.org/10.1016/j.infrared.2016.11.001.
Sahnoun M, Kallel F, Dammak M, Mhiri C, Mahfoudh KB, Hamida AB. A comparative study of MRI contrast enhancement techniques based on Traditional Gamma Correction and Adaptive Gamma Correction: Case of multiple sclerosis pathology. Proceedings of In 2018 4th international conference on advanced technologies for signal and image processing (ATSIP), IEEE. 2018 Mar 21, 1-7. doi: 10.1109/ATSIP.2018.836 4467.
Shreyamsha Kumar BK. Image denoising based on non-local means filter and its method noise thresholding. SIViP. 2013; 7: 1211-27. doi.org/10.1007/s11760-012-0389-y
Chandrashekar L, Sreedevi A. Assessment of nonlinear filters for MRI images. Proceedings of In 2017 2nd International Conference on Electrical, Computer and Communication Technologies (ICECCT), IEEE. 2017 Feb 22, 1-5. doi: 10.1109/ICECCT.2017.8117852.
Min A, Kyu ZM. MRI images enhancement and tumor segmentation for brain. In2017 18th International Conference on Parallel and Distributed Computing, Applications and Technologies (PDCAT) 2017 Dec 18 (pp. 270-275). IEEE. doi: 10.1109/PDCAT.2017.00051.
Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. Proceedings of InMedical Image Computing and Computer-Assisted Intervention-MICCAI 2015: 18th International Conference, Munich, Germany, October 5-9, 2015, 234-241). Springer, Cham. doi.org/10.1007/978-3-319-24574-4_28.
Ghosh S, Santosh KC. Tumor segmentation in brain MRI: U-Nets versus feature pyramid network. Proceedings of In2021 IEEE 34th International Symposium on Computer-Based Medical Systems (CBMS), IEEE. 2021 Jun 7, 31-6. doi: 10.1109/CBMS52027.2021.00013.
Al Nasim MA, Al Munem A, Islam M, Palash MA, Haque MM, Shah FM. Brain tumor segmentation using enhanced u-net model with empirical analysis. In2022 25th International Conference on Computer and Information Technology (ICCIT) 2022 Dec 17 (pp. 1027-32). IEEE. doi: 10.1109/ICCIT57492.2022.10054934.
Manasa K, Krishnaveni V. Brain Tumor Segmentation Using Zernike Moments in U-Net. In 2022 International Conference on Intelligent Innovations in Engineering and Technology (ICIIET) 2022 Sep 22 (pp. 342-6). IEEE. doi: 10.1109/ICIIET55458.2022.9967618.
Yan C, Ding J, Zhang H, Tong K, Hua B, Shi S. SEResU-Net for Multimodal Brain Tumor Segmentation. IEEE Access. 2022; 10: 117033-44. doi: 10.1109/ACCESS.2022.3214309.