CRISPR 2 spacer architecture analysis and virulotyping for epidemiological study of Salmonella enterica subsp. Enterica circulating in northern Thailand (2015 -2017)
Main Article Content
Abstract
Background: Salmonella enterica subsp. enterica, particularly serotype S. 4[5],12:i:-, S. Typhimurium, and S. Enteritidis, represents a significant causative agent of diarrhea, particularly impacting children and immunocompromised individuals on a global scale. Molecular typing of Salmonella spp. has a vital role in understand Salmonella epidemiology.
Objective: The objective of this study is to utilize CRISPR 2 spacer analysis coupled with multiple-locus variable number tandem-repeat (VNTR) analysis and virulotyping to perform molecular typing and potential subtyping of Salmonella spp.
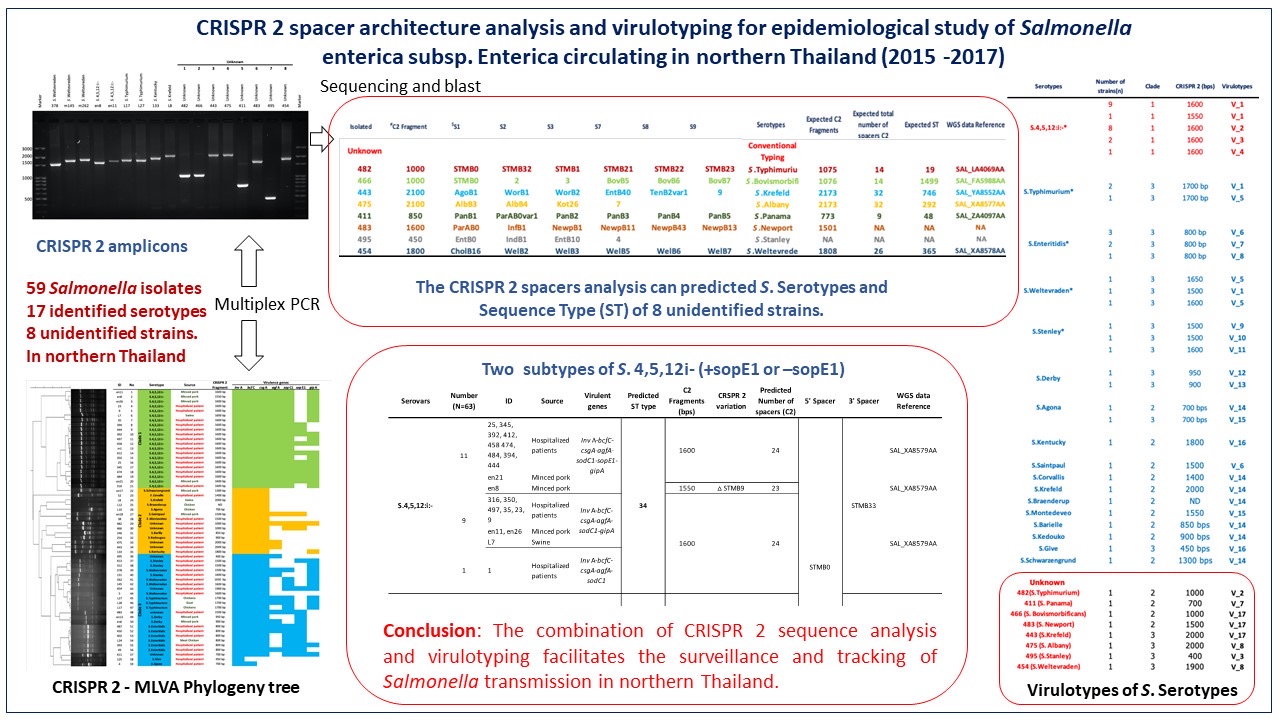
Materials and methods: CRISPR 2 - multiple-locus variable number tandem-repeat (VNTR) analysis, complemented by additional virulotyping, were performed to rapidly characterize those Salmonella isolates including eight unidentified strains. Serotype-specific CRISPR 2 amplicons were subjected to sequencing and the obtained sequences were blasted with corresponding whole-genome sequencing (WGS) data in order to extract CRISPR 2 information, especially the number and sequence of spacers which were then utilized to predict Salmonella serotypes. Moreover, the similar CRISPR 2 spacer architectures to the corresponding WGS offered the prediction of multilocus sequence types (MLST).
Results: S. 4,[5],12:i:-, S. Typhimurium, S. Enteritidis, S. Weltevraden, and S. Derby exhibited distinct clustering, while eight unidentified Salmonella serotypes displayed unique CRISPR 2-MLVA profiles. Through subsequent sequence analysis and comparison with publicly available whole-genome sequencing data, serotype-specific CRISPR 2 amplicon lengths and spacer architectures were unveiled, enabling precise prediction of MLST types. Intriguingly, a linear correlation emerged between CRISPR 2 amplicon length (500-2000 bps) and the number of spacers (6-32) across diverse Salmonella serotypes. Critically, the molecular signatures of CRISPR 2 amplicons accurately predicted the identity of eight unknown Salmonella isolates, aligning with conventional serotyping standards. Furthermore, MLST sequences for prevalent S. 4,[5],12:i:-, S. Typhimurium, and S. Enteritidis were unveiled as ST 34, ST 19, and ST 10, respectively. Subtyping of S. 4,[5],12:i:- using the sopE1 procession (a bacteriophage gene) revealed two major subtypes within ST 34. These subtypes encompassed all six virulent genes, including InvA, bcfC, csgA, agfA, sodC1, and gipA, either with sopE1 (N=8) or without sopE1 (N=10). These findings contribute preliminary insights into the genetic diversity and subtyping of S. 4,[5],12:i:-.
Conclusion: The combination of CRISPR 2 sequence analysis and virulotyping emerged as a potent epidemiological tool, facilitating the identification of Salmonella serotypes and potentially informative subtypes, thereby aiding in the surveillance, and tracking of Salmonella transmission in northern Thailand.
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Personal views expressed by the contributors in their articles are not necessarily those of the Journal of Associated Medical Sciences, Faculty of Associated Medical Sciences, Chiang Mai University.
References
Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, et al. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis. 2010; 50: 882-9.
Kauffmann F. Serological diagnosis of Salmonellaspecies Kauffmann-White-Schema (Scandinavian university books) Vind, Denmark: Munksgaard;1972
Hohmann EL.Nontyphoidal Salmonellosis. Clin Infect Dis. 2001; 32: 263-9.
Chung N, Wang SM, Shen CF, Kuo FC, Ho TS, Hsiung CA, et al. Clinical and epidemiological characteristics in hospitalized young children with acute gastroenteritis in southern Taiwan. J Microbiol Immunol Infect. 2017; 50(6): 915-22. doi: 10.1016/j.jmii.2017.07.015.
Pegues DA, OhH ME, Miller SI. Nontyphoidal Salmonellosis. Trop Infect Dis. 2006; 1: 241-4.
Eng SK, Pusparajah P, Ab Mutalib NS, Ser HL, Chan KG, Lee LH. a. Salmonella: A review on pathogenesis, epidemiology and antibiotic resistance. Front Life Sci. 2015; 8: 284-293.
Cellucci T, Seabrook JA, Chagla Y, Bannister SL, Salvadori MI. A 10-year retrospective review of Salmonella infections at the Children’s Hospital in London, Ontario. Can J Infect Dis Med Microbiol. 2010; 21: 78-82.
De Jong B, Ekdahl K. The comparative burden of salmonellosis in the European Union member states, associated and candidate countries. BMC Public Health. 2006; 6: 1-9.
Centers for Disease Control and Prevention (CDC). 2016. National Enteric Disease Surveillance: Salmonella Annual Report, 2011. Online.
Hörmansdorfer S, Messelhäußer U, Rampp A, Schönberger K, Dallman T, Allerberger F, et al. Re-evaluation of a 2014 multi-country european outbreak of Salmonella enteritidis phage type 14b using recent epidemiological and molecular data. Euro Surveill. 2017; 22: 1-7. doi: 10.2807/1560-7917.ES.2017.22.50.17-00196.
Jourdan-da Silva N, Fabre L, Robinson E, Fournet N, Nisavanh A, Bruyand M, et al. Ongoing nationwide outbreak of Salmonella agona associated with internationally distributed infant milk products, France, December 2017. Euro Surveill. 2018; 23: 1-5. doi: 10.2807/1560-7917.ES.2018.23.2.17-00852.
Bangtrakulnonth A, Pornreongwong S, Pulsrikarn C, Sawanpanyalert P, Hendriksen RS, Lo Fo Wong DM a, et al. Salmonella Serovars from Humans and Other Sources in Thailand, 1993-2002. Emerg Infect Dis. 2004; 10: 131-6. doi: 10.3201/eid1001.02-0781.
Tungwongjulaniam C. Situation of food poisoning, 2017. Dis Control J. 2018; 44(3): 315-24.
ISO 6579:2000. 2002. Microbiology of food and animal feeding stuffs - Horizontal method for the detection of Salmonella spp. Geneva, Switzerland.
Buchan BW, Ledeboer NA. Emerging technologies for the clinical microbiology laboratory. Clin Microbiol Rev. 2014; 27: 783-822. doi: 10.1128/CMR.00003-14.
Wattiau P, Boland C, Bertrand S. Methodologies for Salmonella enterica subsp. Enterica Subtyping: Gold Standards and Alternatives. Appl Environ Microbiol. 2011; 77: 7877-85. doi: 10.1128/AEM.05527-11.
Masek BJ, Hardick J, Won H, Yang S, Hsieh YH, Rothman RE, et al. Sensitive detection and serovar differentiation of typhoidal and nontyphoidal Salmonella enterica species using 16S rRNA gene PCR coupled with high-resolution melt analysis. J Mol Diagn. 2014; 16: 261-6. doi.org/10.1016/j.jmoldx.2013.10.011
Muñoz N, Diaz-Osorio M, Moreno J, Sánchez-Jiménez M, Cardona-Castro N. Development and evaluation of a multiplex real-time polymerase chain reaction procedure to clinically type prevalent Salmonella enterica serovars. J Mol Diagn. 2010; 12: 220-5. doi: 10.2353/jmoldx.2010.090036.
Zeinzinger J, Pietzka AT, Stöger A, Kornschober C, Kunert R, Allerberger F, et al. One-step triplex high-resolution melting analysis for rapid identification and simultaneous subtyping of frequently isolated Salmonella serovars. Appl. Environ. Microbiol. 2012; 78: 3352-60. doi: 10.1128/AEM.07668-11
Leekitcharoenphon P, Nielsen EM, Kaas RS, Lund O, Aarestrup FM. Evaluation of Whole Genome Sequencing for Outbreak Detection of Salmonella enterica. PLoS One. 2014; 4; 9(2): e87991. doi: 10.1371/journal.pone.0087991.
Quick J, Ashton P, Calus S, Chatt C, Gossain S, Hawker J, et al. Rapid draft sequencing and real-time nanopore sequencing in a hospital outbreak of Salmonella. Genome Biol. 2015; 16: 1-14. doi: 10.1186/s13059-015-0677-2.
Jansen R, Van Embden JDA, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002; 43: 1565-75. doi: 10.1046/j.1365-2958.2002.02839.x.
Newsom Sydney, Parameshwaran Hari Priya, Martin Lindsie, Rajan Rakhi. The CRISPR-Cas Mechanism for Adaptive Immunity and Alternate Bacterial Functions Fuels Diverse Biotechnologies. Front Cell Infect Microbiol. 2021; 10: 1-10. doi: 10.3389/fcimb.2020.619763
Shariat N, Dudley EG. CRISPRs: molecular signatures used for pathogen subtyping. Appl Environ Microbiol. 2014; 80(2): 430-9. doi: 10.1128/AEM.02790-13. Epub 2013 Oct 25. PMID: 24162568; PMCID: PMC3911090.
Horvath P, Barrangou R.CRISPR/Cas, the immune system of Bacteria and Archaea. Science. 2010; 327: 167-70. doi: 10.1126/science.1179555.
Fabre L, Zhang J, Guigon G, Le Hello S, Guibert V, Accou-Demartin M, et al. Crispr typing and subtyping for improved Laboratory surveillance of Salmonella infections. PLoS ONE. 2012; 7(5): e36995. doi: 10.1371/journal.pone.0036995. Epub 2012 May 18.
Liu F, Kariyawasam S, Jayarao BM, Barrangou R, GernerSmidt P, Ribot EM, et al. Subtyping Salmonella enterica serovar enteritidis isolates from different sources by using sequence typing based on virulence genes and clustered regularly interspaced short palindromic repeats (CRISPRs). Appl Environ Microbiol. 2011; 77: 4520-6. doi: 10.1128/AEM.00468-11.
Wisittipanit N, Pulsrikarn C, Srisong S, Srimora R, Kittiwan N, Poonchareon K. 2020a. CRISPR 2 PCR and high resolution melting profiling for identification and characterization of clinically-relevant Salmonella enterica subsp. enterica. Peer J. 2020; 8: e9113. doi: 10.7717/peerj.9113.
Thompson CP, Doak AN, Amirani N, Schroeder EA, Wright J, Kariyawasam S, et al. High-resolution identification of multiple Salmonella serovars in a single sample by using CRISPRSeroSeq. Appl Environ Microbiol. 2018; 84: 1-13. doi: 10.1128/AEM.01859-18.
Nadon CA, Trees E, KNg L, Nielsen EM, Reimer A, Maxwell N, et al. Development and application of MLVA methods as a tool for inter-laboratory surveillance. Euro surveill. 2017; 18: 20565. doi: 10.2807/15607917.es2013.18.35.20565.
Wuyts V, Mattheus W, De Laminne De Bex G, Wildemauwe C, Roosens NHC, et al. MLVA as a tool for public health surveillance of human Salmonella Typhimurium: Prospective study in Belgium and evaluation of MLVA loci stability. PLoS ONE. 2013; 8(12): e84055. doi.org/10.1371/journal.pone.0084055
Caméléna F, Birgy A, Smail Y, Courroux C, MarianiKurkdjian P, Hello S Le, et al. Rapid and Simple Universal Escherichia coli Genotyping Method Based on MultipleLocus Variable-Number Tandem- Repeat Analysis Using Single-Tube Multiplex PCR and Standard Gel Electrophoresis. Appl Environ Microbiol. 2019; 85: 1-15.
Huehn S, La Ragione RM, Anjum M, Saunders M, Woodward MJ, Bunge C, et al. Virulotyping and antimicrobial resistance typing of salmonella enterica serovars relevant to human health in Europe. Foodborne Pathog Dis. 2010; 7: 523-35. doi: 10.1089/fpd.2009.0447.
Drahovská H, Mikasová E, Szemes T, Ficek A, Sásik M, Majtán V, et al.Variability in occurrence of multiple prophage genes in Salmonella Typhimurium strains isolated in Slovak Republic. FEMS Microbiol Lett. 2007; 270: 237-44. doi: 10.1111/j.1574-6968.2007.00674.x.
Poonchareon K, Pulsrikarn C, Khamvichai S, Tadee P. Feasibility of high resolution melting curve analysis for rapid serotyping of Salmonella from hospitalised patients. J Assoc Med Sci. 2019; 52(1): 36-40. doi: 10.14456/jams.2018.3.
Poonchareon K, Pulsrikarn C, Nuanmuang N, Khamai P.
Effectiveness of BOX-PCR in Differentiating Genetic Relatedness among Salmonella enterica Serotype 4,[5],12:i:- Isolates from Hospitalized Patients and Minced Pork Samples in Northern Thailand. Int J Microbiol. 2019; 2019: 5086240. doi: 10.1155/2019/5086240.
Borriello G, Lucibelli MG, Pesciaroli M, Carullo MR, Graziani C, Ammendola S, et al. Diversity of Salmonella spp. serovars isolated from the intestines of water buffalo calves with gastroenteritis. BMC Vet Res. 2012; 8: 201. doi: 10.1186/1746-6148-8-201.
Rahn K, Grandis A De, Clarke RC, McEwen S. A, Galin JE, Ginocchio C, et al. Amplification of invA gene of Salmonella by polymerase chain reaction (PCR) as a specific method for detection of Salmonellae. Mol Cell Probes. 1992; 6: 271-9. doi: 10.1016/08908508(92)90002-f
McNerney R, Clark TG, Campino S, Rodrigues C, Dolinger
D, Smith L, et al.Removing the bottleneck in whole genome sequencing of Mycobacterium tuberculosis for rapid drug resistance analysis: a call to action. Int J Infect Dis. 2017; 56: 130-5. doi: 10.1016/j.ijid.2016.11.422.
Heras, J., Domínguez, C., Mata, E. et al. GelJ - a tool for analyzing DNA fingerprint gel images. BMC Bioinformatics 2015; 16: 270. doi.org/10.1186/s12859-015-0703-0
Eng S-KK, Pusparajah P, Ab Mutalib N-SS, Ser H-LL, Chan K-GG, Lee L-HH. Salmonella: A review on pathogenesis, epidemiology and antibiotic resistance. Front Life Sci. 2015; 8: 284-93. doi: 10.1080/21553769.2015.1051243.
Gordon MA, Graham SM, Walsh AL, Wilson L, Phiri A, Molyneux E, et al. Epidemics of invasive Salmonella enterica serovar enteritidis and S. enterica serovar typhimurium infection associated with multidrug resistance among adults and children in Malawi. Clin Infect Dis. 2008; 46: 963-9. doi: 10.1086/529146.
Elbagir M, Nori E, Thong KL. Differentiation of Salmonella enterica based on PCR detection of selected somatic and flagellar antigens. Afr J Microbiol Res. 2010; 4: 871-6. doi: 10.5897/AJMR.9000238.
Poonchareon K, Narong Nuanmuang, Prommuang P, Sriisan S. High-resolution melting-curve analysis for serotyping of Salmonella spp. group B isolated from minced pork in the Northern part of Thailand. J Assoc Med Sci. 2019; 52(1): 62-71. doi: 10.14456/jams.2018.3.
Wisittipanit N, Pulsrikarn C, Wutthiosot S, Pinmongkhonkul S, Poonchareon K. Application of machine learning algorithm and modified high resolution DNA melting curve analysis for molecular subtyping of Salmonella isolates from various epidemiological backgrounds in northern Thailand. World J Microbiol Biotechnol. 2020; 36: 103. doi: 10.1007/s11274-020-02874-7.
Lindstedt BA, Heir E, Nygård I, Kapperud G. Characterization of class I integrons in clinical strains of Salmonella enterica subsp. enterica serovars Typhimurium and Enteritidis from Norwegian hospitals. J Med Microbiol. 2003; 52: 141-9. doi: 10.1099/jmm.0.04958-0.
Mandilara G, Lambiri M, Polemis M, Passiotou M, Vatopoulos A. Phenotypic and molecular characterisation of multiresistant monophasic Salmonella typhimurium (1,4,[5],12:I:-) in Greece, 2006 to 2011. Euro Surveill. 2013; 18: 1-8.
Xie X, Wang Z, Zhang K, Li Y, Hu Y, Pan Z, et al. Pig as a reservoir of CRISPR type TST4 Salmonella enterica serovar Typhimurium monophasic variant during 2009- 2017 in China. Emerg Microbes Infect. 2020; 9: 1-4. doi: 10.1080/22221751.2019.1699450.
Yang X, Wu Q, Zhang J, Huang J, Guo W, Cai S. Prevalence and characterization of monophasic Salmonella serovar 1,4,[5],12:i:-of food origin in China. PLoS ONE. 2015; 10: 1-10. doi: 10.1371/journal.pone.0137967.
Zhang S, Santos RL, Tsolis RM, Mirold S, Hardt WD, Adams LG, et al. Phage mediated horizontal transfer of the sopE1 gene increases enteropathogenicity of Salmonella enterica serotype Typhimurium for
calves. FEMS Microbiol Lett. 2002; 217: 243-7. doi: 10.1016/S0378-1097(02)01094-7.
Tanmoy AM, Saha C, Sajib MSI, Saha S, KomurianPradel F, Belkum A van, et al. CRISPR-Cas Diversity in Clinical Salmonella enterica Serovar Typhi Isolates from South Asian Countries. Genes (Basel). 2020; 11(11): 1365. doi: 10.3390/genes11111365. PMID: 33218076; PMCID: PMC7698835.