Evaluation of conventional methods for species identification of Staphylococcus aureus using MALDI-TOF MS, protein identification and quantification
Main Article Content
Abstract
Objectives: This study aimed to evaluate conventional methods for species identification of Staphylococcus aureus by MALDI-TOF MS. Additionally, the representatives of different species were used to analyze protein expression.
Materials and methods: A total of 185 non-duplicated clinical S. aureus identified using the conventional method (colony morphology, Gram’s stain, slide coagulase test, tube coagulase test, catalase test, and mannitol fermentation) was confirmed the identification by MALDI-TOF MS and analyzed by Mass spectral profiling (MSP) and Principal component analysis (PCA). The representatives of different species reported by both methods were confirmed using 16SrRNA sequence and analyzed proteins expression by timsTOF MS.
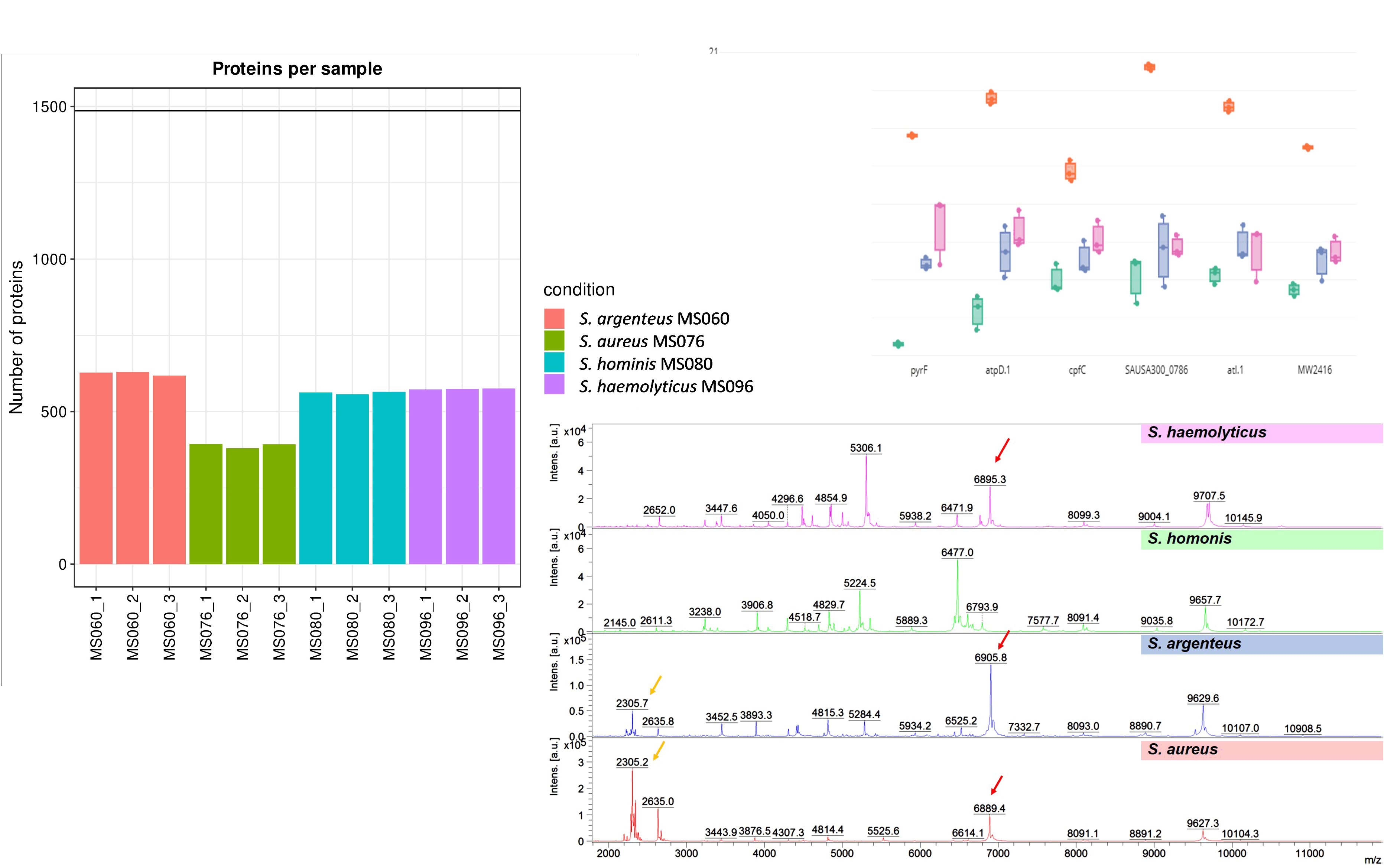
Results: All S. aureus suspected isolates could discriminate among species by MALDI-TOF MS including S. aureus (N=151, 81.6%), S. argenteus (N=32, 17.3%), S. hominis (N=1, 0.5%), and S. haemolyticus (N=1, 0.5%). Using 16S rRNA genebased analysis, S. aureus and S. argenteus could not differentiate from each other. Protein expression of S. aureus was similar to S. argenteus. These genes including rpsT, Huti, pyrF, atpD.1, cpfC, SAUA300_0786, atl.1, and MW2416 showed higher expression in S. aureus (MS076) than S. argenteus (MS060), S. haemolyticus (MS095) and S. hominis (MS060).
Conclusion: MALDI-TOF MS provides an excellent tool for accurately species identification of staphylococci. S. aureus expressed protein analyzed higher than the other 3 species. The highest protein expression in S. aureus implies the most virulence of this strain.
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Personal views expressed by the contributors in their articles are not necessarily those of the Journal of Associated Medical Sciences, Faculty of Associated Medical Sciences, Chiang Mai University.
References
Mehta Y, Hegde A, Pande R, Zirpe KG, Gupta V, Ahdal J, Qamra A, Motlekar S, Jain R. Methicillin-resistant Staphylococcus aureus in Intensive Care Unit Setting of India: A Review of Clinical Burden, Patterns of Prevalence, Preventive Measures, and Future Strategies. Indian J Crit Care Med. 2020; 24(1): 55- 62. doi: 10.5005/jp-journals-10071-23337.
Onyango LA, Alreshidi MM. Adaptive Metabolism in Staphylococci: Survival and Persistence in Environmental and Clinical Settings. J Pathog. 2018; 2018: 1092632.
doi.org/10.1155/2018/1092632.
Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG, Jr. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015; 28(3): 603-61. doi: 10.1128/ CMR.00134-14.
Ayeni FA, Andersen C, Nørskov-Lauritsen N. Comparison of growth on mannitol salt agar, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, VITEK 2 with partial sequencing of 16S rRNA gene for identification of coagulase-negative staphylococci. Microb. Pathog. 2017; 105: 255-9. doi: 10.1016/j. micpath.2017.02.034.
Zhang L, Sandrin TR. Maximizing the Taxonomic Resolution of MALDI-TOF-MS-Based Approaches to Bacterial Characterization: From Culture Conditions Through Data Analysis. 2016: 147-81. doi: 10.1007/ 978-3-319-26070-9_6.
Singhal N, Kumar M, Kanaujia PK, Virdi JS. MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front Microbiol. 2015; 6: 791-6. doi: 10.3389/fmicb.2015.00791.
Kim E, Kim HJ, Yang SM, Kim CG, Choo DW, Kim HY. Rapid identification of Staphylococcus species isolated from food samples by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Microbiol Biotechnol. 2019; 29: 548-57. doi: 10.4014/ jmb.1901.01046.
Shao C, Tian Y, Dong Z, Gao J, Gao Y, Jia X, et al. The use of principal component analysis in MALDI-TOF MS: a powerful tool for establishing a mini-optimized proteomic profile. Am J Biomed Sci. 2012; 4(1): 85- 101. doi: 10.5099/aj120100085.
Wang H-Y, Lee T-Y, Tseng Y-J, Liu T-P, Huang K-Y, Chang Y-T, et al. A new scheme for strain typing of methicillin-resistant Staphylococcus aureus on the basis of matrix-assisted laser desorption ionization time-of-flight mass spectrometry by using machine learning approach. PLoS One. 2018; 13(3): e0194289. doi: 10.1371/journal.pone.0194289.
Thitiya Yungyuen, Tanittha Chatsuwan, Rongpong Plongla, Sakawrat Kanthawong, Umaporn Yordpratum, Supayang P. Voravuthikunchai, et al. Nationwide surveillance and molecular characterization of critically drug-resistant Gram-negative bacteria: Results of the research university network Thailand study. Antimicrob Agent Chemother 2021; 65(9): e00675-21.doi:10.1128 /aac.00675-21.
Chuanboon K, Na Nakorn P, Pannengpetch S, Laengsri V, Nuchnoi P, Isarankura-Na-Ayudhya C, IsarankuraNa-Ayudhya P. Proteomics and bioinformatics analysis reveal potential roles of cadmium-binding proteins in cadmium tolerance and accumulation of Enterobacter cloacae. Peer J. 2019;7: e6904. doi: 10.7717/ peerj.6904.
Na Nakorn P, Pannengpetch S, Isarankura-Na-Ayudhya P, Thippakorn C, Lawung R, Sathirapongsasuti N, Kitiyakara C, Sritara P, Vathesatogkit P, Isarankura-NaAyudhya C. Roles of kininogen-1, basement membrane specific heparan sulfate proteoglycan core protein, and roundabout homolog 4 as potential urinary protein biomarkers in diabetic nephropathy. EXCLI J. 2020; 19: 872-91. doi: 10.17179/excli2020-1396.
Meier F, Beck S, Grassl N, Lubeck M, Park MA, Raether O, Mann M. Parallel accumulation-serial fragmentation (PASEF): multiplying sequencing speed and sensitivity by synchronized scans in a tapped ion mobility device. J Proteome Res. 2015; 14(12): 5378- 87. doi: 10.1021/acs.jproteome.5b00932.
Yu F, Haynes SE, Teo GC, Avtonomov DM, Polasky DA, Nesvizhskii AI. Fast quantitative analysis of timsTOF PASEF data with MSFragger and IonQuant. Mol Cell Proteomics. 2020; 19(9): 1575-85. doi: 10.1074/mcp.
TIR120.002048.
Kong AT, Leprevost FV, Avtonomov DM, Mellacheruvu D, Nesvizhskii AI. MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat Methods. 2017; 14(5): 513-20. doi: 10.1038/nmeth.4256.
Thakur P, Nayyar C, Tak V, Saigal K. Mannitol-fermenting and tube coagulase-negative staphylococcal isolates: unraveling the diagnostic dilemma. J Lab Physicians. 2017; 9(1): 65-6. doi: 10.4103/0974-2727.187926.
Chantratita N, Wikraiphat C, Tandhavanant S, Wongsuvan G, Ariyaprasert P, Suntornsut P, et al. Comparison of community-onset Staphylococcus argenteus and Staphylococcus aureussepsis in Thailand: a prospective multicentre observational study. Clin Microbiol Infect. 2016; 22(5): 458.e11-9. doi: 10.1016/j.cmi. 2016.01.008.
Hansen TA, Bartels MD, Høgh SV, Dons LE, Pedersen M, Jensen TG, et al. Whole genome sequencing of Danish Staphylococcus argenteus reveals a genetically diverse collection with clear separation from Staphylococcus aureus. Front Microbiol. 2017; 8 (1512). doi: 10.3389/fmicb.2017.01512.
Kosecka-Strojek M, Sabat AJ, Akkerboom V, Becker K, van Zanten E, Wisselink G, Miedzobrodzki J, KooistraSmid AMDM, Friedrich AW. Development and validation of a reference data set for assigning Staphylococcus species based on next-generation sequencing of the 16S-23S rRNA region. Front Cell Infect Microbiol. 2019; 9: 278. doi: 10.3389/ fcimb.2019.00278.
Schuster D, Rickmeyer J, Gajdiss M, Thye T, Lorenzen S, Reif M, Josten M, Szekat C, Melo LDR, Schmithausen RM, Liégeois F, Sahl HG, Gonzalez JJ, Nagel M, Bierbaum G. Differentiation of Staphylococcus argenteus (formerly: Staphylococcus aureus clonal complex 75) by mass spectrometry from S. aureus using the first strain isolated from a wild African great ape. Int J Med Microbiol. 2017; 307(1): 57-63. doi: 10.1016/j. ijmm.2016.11.003.
Liu Z, Wang H, Zhou Z, Naseer N, Xiang F, Kan B, Goulian M, Zhu J. Differential Thiol-Based Switches JumpStart Vibrio cholerae Pathogenesis. Cell Rep. 2016; 14: 347-54. doi: 10.1016/j.celrep.2015.12.038.
Pande A, Veale TC, Grove A. Gene regulation by redoxsensitive Burkholderia thailandensis OhrR and its role in bacterial killing of Caenorhabditis elegans. Infect Immun. 2018; 86(9): e00322-18. doi: 10.1128/IAI. 00322-18.
Sun M, Lyu M, Wen Y, Song Y, Li J, Chen Z. Organic peroxide-sensing repressor OhrR regulates organic hydroperoxide stress resistance and avermectin production in Streptomyces avermitilis. Front Microbiol. 2018; 9:1398. doi: 10.3389/fmicb.2018.01398.
Bosch ME, Bertrand BP, Heim CE, Alqarzaee AA, Chaudhari SS, Aldrich AL, Fey PD, Thomas VC, Kielian T. Staphylococcus aureusATP synthase promotes biofilm persistence by influencing innate immunity. mBio. 2020; 11(5): e01581-20. doi: 10.1128/mBio.01581-20.
Porayath C, Suresh MK, Biswas R, Nair BG, Mishra N, Pal S. Autolysin mediated adherence of Staphylococcus aureus with fibronectin, gelatin, and heparin. Int J Biol Macromol. 2018; 110: 179-84. doi: 10.1016/j. ijbiomac.2018.01.047.
Lei MG, Cue D, Roux CM, Dunman PM, Lee CY. Rsp inhibits attachment and biofilm formation by repressing fnbA in Staphylococcus aureus MW2. J Bacteriol. 2011; 193(19): 5231-41. doi: 10.1128/JB. 05454-11.
Carrera-Salinas A, González-Díaz A, Vázquez-Sánchez DA, Camoez M, Niubó J, Càmara J, Ardanuy C, Martí S, Domínguez MÁ; REIPI/GEIH Study Groups. Staphylococcus aureus surface protein G (sasG) allelic variants: correlation between biofilm formation and their prevalence in methicillin-resistant S. aureus (MRSA) clones. Res Microbiol. 2022; 173(3): 103921. doi: 10.1016/j.resmic.2022.103921.
Shibamura-Fujiogi M, Wang X, Maisat W, Koutsogiannaki S, Li Y, Chen Y, et al. GltS regulates biofilm formation in methicillin-resistant Staphylococcus aureus. Commun Biol. 2022; 5(1): 1284. doi: 10.1038/s42003-022-042 39-2.
Hobbs C, Reid JD, Shepherd M. The coproporphyrin ferrochelatase of Staphylococcus aureus: mechanistic insights into a regulatory iron-binding site. Biochem J.
; 474(20): 3513-22. doi: 10.1042/BCJ20170362.
Kim GL, Hooven TA, Norambuena J, Li B, Boyd JM, Yang JH, Parker D. Growth and stress tolerance comprise independent metabolic strategies critical for Staphylococcus aureus infection. mBio. 2021; 12(3): e0081421. doi: 10.1128/mBio.00814-21.
Knöppel A, Andersson DI, Näsvall J. Synonymous mutations in rpsT lead to ribosomal assembly defects that can be compensated by mutations in fis and rpoA. Front Microbiol. 2020; 11: 340. doi: 10.3389/fmicb.2020.00340.
Eshaghi A, Bommersbach C, Zittermann S, Burnham CA, Patel R, Schuetz AN, Patel SN, Kus JV. Phenotypic and genomic profiling of Staphylococcus argenteus in Canada and the United States and recommendations for clinical result reporting. J Clin Microbiol. 2021; 59(6): e02470-20. doi: 10.1128/JCM.02470-20.
Wayne PA, Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. CLSI M100, 32nd Ed. Clinical and Laboratory Standards Institute. 2022.
Tsuchida S, Umemura H, Nakayama T. Current status of matrix-assisted laser desorption/ionization-time-offlight mass spectrometry (MALDI-TOF MS) in clinical diagnostic microbiology. Molecules. 2020; 25(20):
doi: 10.3390/molecules25204775.