Evaluation of novel PCR-CTPP for simultaneous detection of Mycobacterium tuberculosis complex and identification of RpoB H526D point mutation
Main Article Content
Abstract
Background: Tuberculosis (TB) is a chronic and highly contagious disease caused of the acid-fast bacilli, Mycobacterium tuberculosis complex (MTC). Currently, it has been reported that rifampicin-mono resistant M. tuberculosis containing histidine to aspartate replacement at residue 526 (H526D) of the beta-subunit RNA polymerase enzyme (RpoB) increased the cell wall permeability and approximately eight times more susceptible to vancomycin.
Objectives: To develop and evaluate the Polymerase chain reaction with confronting two-pair primers (PCR-CTPP) for simultaneous detection of MTC and identification of RpoB H526D mutation.
Materials and methods: PCR-CTPP was implemented for TB diagnosis. Reaction and profile were optimized and applied for detection in a total of 308 clinical samples. Sensitivity and specificity of PCR-CTPP was calculated in comparison to the acid-fast bacilli (AFB) staining and standard culture method. In addition, microscopic observation drug susceptibility (MODS) assay was modified for vancomycin susceptibility determination in 15 clinical isolates.
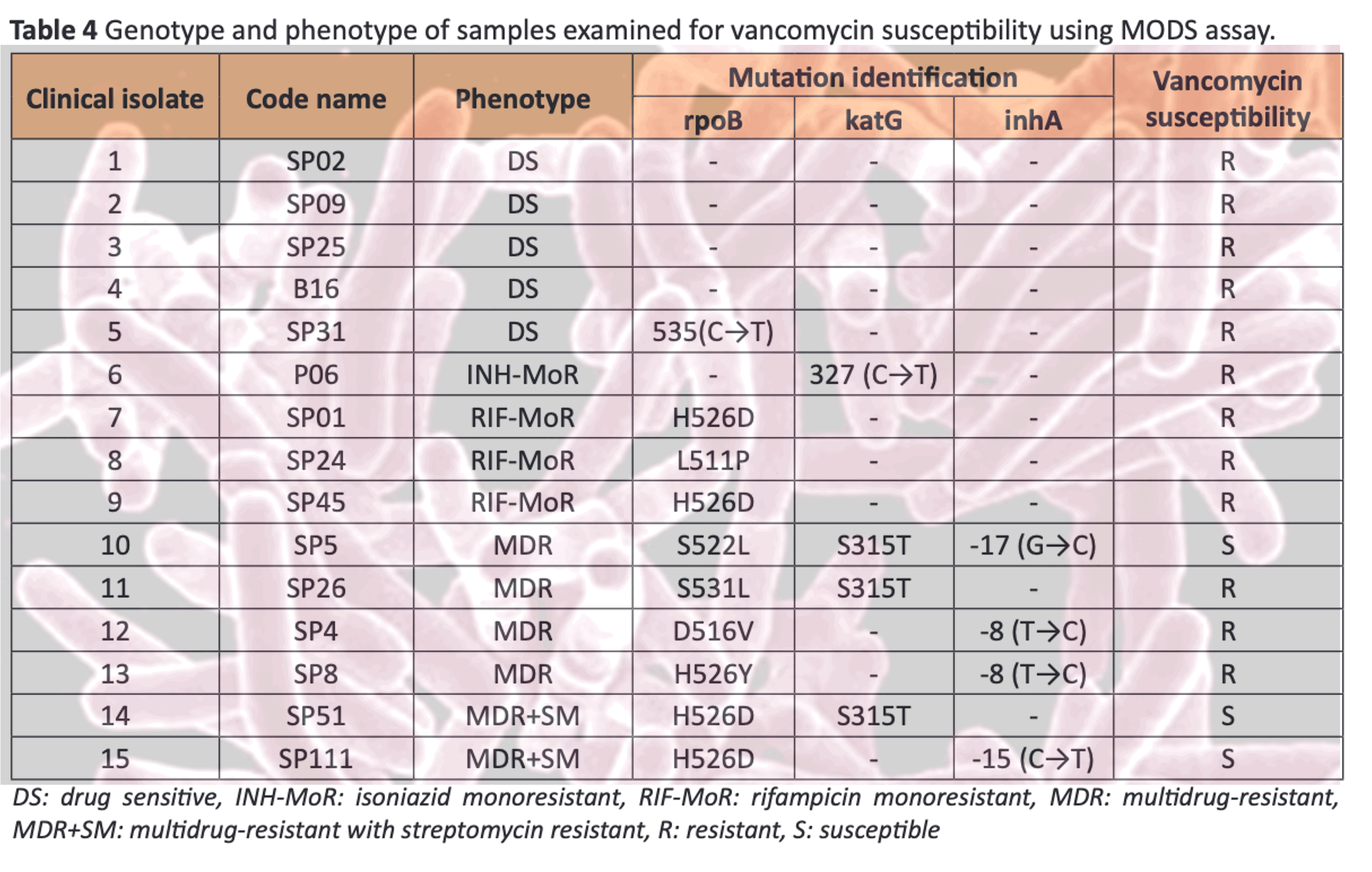
Results: Limit of detection of PCR-CTPP was approximately 2×103 bacilli and no cross-detection to other mycobacteria. PCR-CTPP was evaluated in 308 clinical samples. Sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) of PCR-CTPP versus standard culture method were 90.95%, 79.31%, 91.78% and 77.53%, respectively. In addition, comparison of developed PCR-CTPP versus AFB staining were also represented. MODS was performed in fifteen samples, two multidrug-resistant (MDR) strains containing RpoB H526D were susceptible against vancomycin.
Conclusion: The established PCR-CTPP is highly sensitive, specific for investigation of MTC and identification of RpoB H526D mutation. This method could be useful for TB diagnosis together with precision medicine application in vancomycin susceptibility determination in TB patients in the future.
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Personal views expressed by the contributors in their articles are not necessarily those of the Journal of Associated Medical Sciences, Faculty of Associated Medical Sciences, Chiang Mai University.
References
Cohen A, Mathiasen VD, Schon T, Wejse C. The global prevalence of latent tuberculosis: a systematic review and meta-analysis. Eur Resir J 2019; 54 (3): 1900655. doi: 10.1183/13993003.00655-2019.
World Health Organization. Global tuberculosis report 2020. World Health Organization. 2020. www.who.int/tb/global-report-2020.
Pontali E, Reviglione M, Migliori GB, the writing group members of the global TB network clinical trial committee. Regimens to treat multidrug-resistant tuberculosis: past, present and future perspectives. Eur Respir Rev 2019; 28 (152): 190035. doi: 10.1183/16000617.0035-2019.
Suthum K, Samosornsuk W, Samosornsuk S. Characterization of katG, inhA, rpoB and pncA in Mycobacterium tuberculosis isolates from MDR-TB risk patients in Thailand. J Infect Dev Ctries 2020; 14(3): 268-76. doi: 10.3855/jidc.11974.
Ahmad S, Mokaddas E, Fares E. Characterization of rpoB mutations in rifampin-resistant clinical Mycobacterium tuberculosis isolates from Kuwait and Dubai. Diagn Microbiol Infect Dis 2002; 44 (3): 245-52. doi: 10.1016/s0732-8893(02)00457-1.
Zaw MT, Emran NA, Lin Z. Mutations inside rifampicin-resistance determining region of rpoB gene associated with rifampicin-resistance in Mycobacterium tuberculosis. J Infect Public Health 2018; 11 (5): 605-10. doi: 10.1016/j.jiph.2018.04.005.
Campodónico VL, Rifat D, Chuang YM, Ioerger TR, Karakousis PC. Altered Mycobacterium tuberculosis cell wall metabolism and physiology associated with rpoB mutation H526D. Front Microbiol 2018; 9: 494. doi: 10.3389/fmicb.2018.00494.
Hamajima N. PCR-CTPP: a new genotyping technique in the era of genetic epidemiology. Expert Rev Mol Diagn 2001; 1 (1): 119-23. doi: 10.1586/14737159.1.1.119.
Tharinjaroen CS, Intorasoot S, Anukool U, Phunpae P, Butr-Indr B, Orrapin S, et al. Novel targeting of the lepB gene using PCR with confronting two-pair primers for simultaneous detection of Mycobacterium tuberculosis complex and Mycobacterium bovis. J Med Microbiol 2016; 65 (1): 36-43. doi: 10.1099/jmm.0.000188.
Intorasoot S, Tharinjaroen CS, Phunpae P, Butr-Indr B, Anukool U, Intachai K, et al. Novel potential diagnostic test for Mycobacterium tuberculosis complex using combined immunomagnetic separation (IMS) and PCR-CTPP. J Appl Microbiol 2016; 121 (2): 528-538. doi: 10.1111/jam.13157.
Agarwal A, Katoch CDS, Kumar M, Dhole TN, Sharma YK. Evaluation of microscopic observation drug susceptibility (MODS) assay as a rapid, sensitive and inexpensive test for detection of tuberculosis and multidrug resistant tuberculosis. Med J Armed Forces India 2019; 75 (1): 58-64. doi: 10.1016/j.mjafi.2018.03.011.
Chaiyasirinroje B, Aung MN, Moolphate S, Kasetjaroen Y, Rienthong S, Rienthong D, et al. Prospective evaluation of simply modified MODS assay: an effective tool for TB diagnosis and detection of MDRTB. Infect Drug Resist 2012; 5: 79-86. doi: 10.2147/IDR.S24295.
Acharya S, Ghimire P, Khadka DK, Nepali S. Comparison of proportion and resistance ratio methods for drug susceptibility testing of Mycobacterium tuberculosis isolated from patients attending national tuberculosis centre, Nepal. SAARC J Tuber Lung Dis HIV/AIDS 2008; V (1): 1-8.
Rifat D, Campodonico VL, Tao J, Miller JA, Alp A, Yao Y, et al. In vitro and in vivo fitness costs associated with Mycobacterium tuberculosis RpoB mutation H526D. Future Microbiol 2017; 12 (9): 753-65. doi: 10.2217/fmb-2017-0022.
Manca C, Tsenova L, Barry CE, Bergtold A, Freeman S, Haslett PA, et al. Mycobacterium tuberculosis CD1551 induces a more vigorous host response in vivo and in vitro but is not more virulent than other clinical isolates. J Immunol 1999; 162 (11): 6740-6.
Chakravorty S, Simmons AM, Rowneki M, Parmar H, Cao Y, Ryan J, et al. The new Xpert MTB/RIF Ultra: Improving detection of Mycobacterium tuberculosis and resistance to rifampin in an assay suitable for point-of-care testing. MBio 2017; 8: e00812–7. doi: 10.1128/ mBio.00812-17.
Wang WH, Takeuchi R, Jain SH, Jiang YH, Watanuki S, Ohtaki Y, et al. A novel, rapid (within hours) culture-free diagnostic method for detecting live Mycobacterium tuberculosis with high sensitivity. EbioMedicine 2020; 20: 103007.
Landis RJ & Koch GG. The measurement of observer agreement for categorical data. Biometrics 1977; 33: 159-74.
Dzodanu EG, Afrifa J, Acheampong DO, Dadzie I. Diagnostic yield of fluorescence and ziehl-Neelsen staining techniques in the diagnosis of pulmonary tuberculosis: A comparative study in a district health facility. Tuberc Res Treat 2019; 2019: Article ID 4091937, 6 pages. doi: 10.1155/2019/4091937.
Soejima T, Iida K, Qin T, Taniai H, Seki M, Yoshida S. Method to detect only live bacteria during PCR amplification. J Clin Microbiol 2008; 46 (7): 2305-13. Doi: 10.1128/JCM.02171-07.
Lu J, Zheng H, Chu P, Han S, Yang H, Wang Z, et al. Direct detection from clinical sputum samples to differentiate live and dead Mycobacterium tuberculosis. J Clin Lab Anal 2019; 33: e22716. doi: 10.1002/jcla.22716.
Ekrami A, Samarbaf-Zadeh AR, Khosravi A, Zargar B, Alavi M, Amin M, et al. Validity of bioconjugated silica nanoparticles in comparison with direct smear, culture, and polymerase chain reaction for detection of Mycobacterium tuberculosis in sputum specimens. Int J Nanomed 2011; 6: 2729-35. doi: 10.2147/ijn. s23239.
Mphande-Nyasulu F, Puengpipattrakul P, Praipruksaphan M, Keeree A, Ruanngean K. Prevalence of tuberculosis (TB), including multi-drug-resistant and extensivelydrug-resistant TB, and association with occupation in adults at Sirindhorn Hospital, Bangkok. IJID Regions. 2022; 2: 141-8.
Prammananan T, Cheunoy W, Taechamahapun D, Yorsangsukkamol J, Phunpruch S, Phdarat P, et al. Distribution of rpoB mutations among multidrugresistant Mycobacterium tuberculosis (MDRTB) strains from Thailand and development of a rapid method for mutation detection. Clin Microbiol Infect 2008; 14 (5): 446-53. doi: 10.1111/j.1469-0691.2008.01951.x.
Anukool U, Phunpae P, Sitthidet CT, Butr-Indr B, Saikaew S, Netirat N, et al. Genotypic distribution and a potential diagnostic assay of multidrug-resistant tuberculosis in northern Thailand. Infect Drug Resist 2020; 13: 3375-82.